Cenik Lab
@CenikLab
Followers
443
Following
610
Media
11
Statuses
156
Austin, TX
Joined October 2019
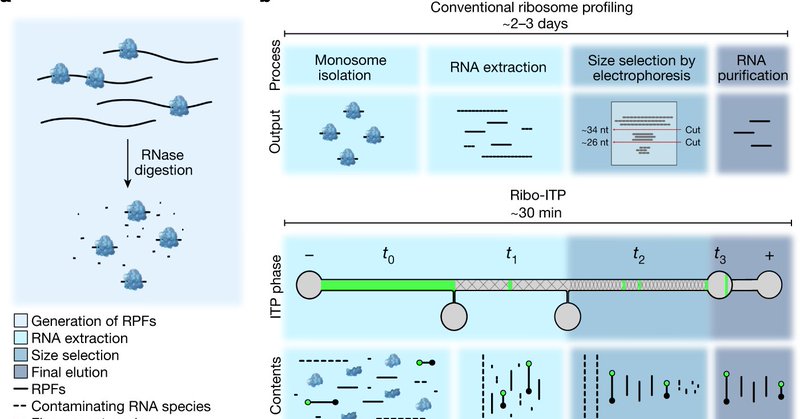
Ribo-ITP enables ribosome occupancy measurements from single cells. Our first application revealed insights into allele-specific ribosome engagement in early development ( https://t.co/BA27Bc5ZGh).
#RiboITP #singlecell
nature.com
Nature - A single-cell ribosome profiling method can provide data at the level of allele-specific ribosome engagement in early development.
9
56
195
@arjunrajlab Earlier this summer I came across this and recommended to a new student in the lab:
sciencecommunicationlab.org
Plan your science career with our FREE course on strategic planning, crafting research questions, and mentorship for students and postdocs.
0
2
2
In our new preprint, we integrated three multiplexed assays of variant effect to profile several layers of gene expression for thousands of variants in COMT!
biorxiv.org
Multiplexed assays of variant effect are powerful methods to profile the consequences of rare variants on gene expression and organismal fitness. Yet, few studies have integrated several multiplexed...
0
5
23
Intrigued by the potential of Ribo-ITP in uncovering the intricacies of cellular translation? We're seeking motivated postdocs to join our team, we'd love to connect with you!
0
2
3
Through collaborations, we're applying Ribo-ITP to diverse areas like early mouse limb development, brain organoids, immune cells, and rare cancer cells. If you are interested in adopting Ribo-ITP, here is our step-by-step protocol with video instructions:
ceniklab.github.io
Translation Control and Computational Biology
1
3
12
Since our preprint, we've delved deeper into translational control of genes that display allele-biased expression. Our findings hint at reduced translation efficiency of such transcripts.
1
0
1
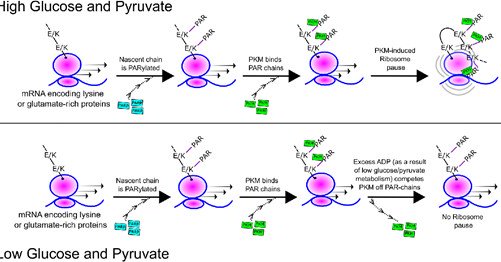
We are happy to unveil our latest published work where we describe how the metabolic enzyme, Pyruvate Kinase (PKM), regulates global mRNA translation in human cells. Here's a short 🧵...
academic.oup.com
Abstract. In light of the numerous studies identifying post-transcriptional regulators on the surface of the endoplasmic reticulum (ER), we asked whether t
5
22
103
Our study showing that Hedgehog signaling is essential for maintaining the early foregut and larynx by preventing EMT was published: https://t.co/6376OAi5dm
@eLife. This was led by recent @UTexasILS graduate Janani Ramachandran with help from Hongkai Ji @jihk99 and @ZornLab.
elifesciences.org
HH signaling prevents precocious EMT and cell death to maintain epithelial cell states during early stages of larynx development.
3
6
32
Congratulations to the 2022 @UT_iGEM team for winning the the Best Foundational Advance award at the iGEM Grand Jamboree! They were also a top 10 overall undergraduate team, finalists for Best Presentation and Best Part Collection, and awarded a Gold Medal. @ut_mbs @TexasScience
0
8
28
Our 1st effort into dissecting the crosstalk between #autophagy & other vacuolar trafficking pathways, now out in @JCellBiol Thx to everyone involved!
‼️ #JustPublished in @JCellBiol New work from the Dagdas Lab at GMI @PlantoPhagy sheds light on the mechanism of #autophagosome maturation in #plants 🌱🌿 Read more: ➡️ https://t.co/HlEy6UX2nZ
#autophagy #plantautophagy #plantorganelle #organelletrafficking #amphisome 1/3
17
49
224
Hypodermal ribosome synthesis inhibition induces a nutrition-uncoupled organism-wide growth quiescence in C. elegans https://t.co/OgHUEFypP9
@SarinayElif
biorxiv.org
Inter-organ communication is a key aspect of multicellular organismal growth, development, and homeostasis. Importantly, cell-non-autonomous inhibitory cues that limit tissue specific growth altera...
0
3
16
It was our pleasure to host @SarinayElif for a virtual seminar today. Thanks a lot Elif for showing us the wonders of C.elegans and sharing your most recent findings with us.
1
1
8
We had a great time at #RNA2022. Lots of exciting talks including from @SarinayElif and @Jamie_Yelland. Looking forward to watching the recorded talks that we missed during the meeting.
0
1
34
Our latest work on HIV-1 translation: Extensive uORF translation from HIV-1 transcripts conditions DDX3 dependency for expression of main ORFs and elicits specific T cell immune responses in infected individuals
biorxiv.org
Human immunodeficiency virus type-1 (HIV-1) is a complex retrovirus which relies on alternative splicing, translational and post-translational mechanisms to produce more than 15 functional proteins...
1
5
36