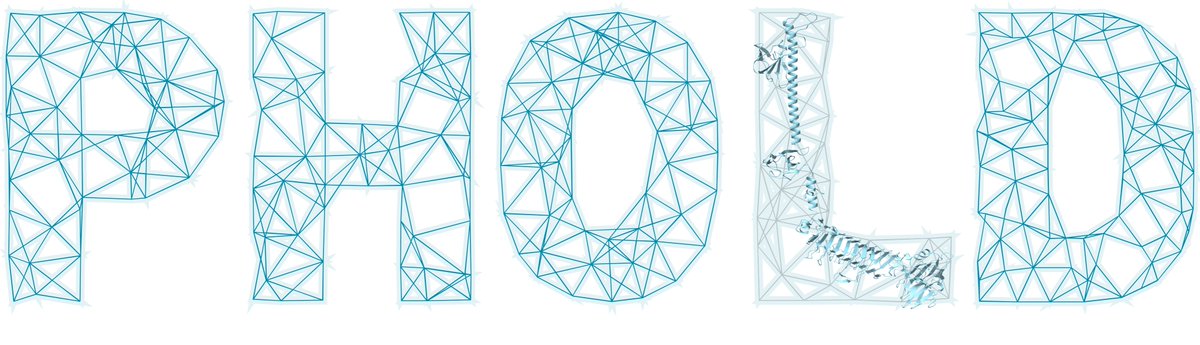Zachary Flamholz
@zflam94
Followers
69
Following
349
Media
4
Statuses
130
current student, future physician scientist | bringing data to medicine | opinions are my own | RT ≠ endorsement
New York, NY
Joined April 2017
AI the scientist knows phages are where it’s at.
biorxiv.org
AI models have been proposed for hypothesis generation, but testing their ability to drive high-impact research is challenging, since an AI-generated hypothesis can take decades to validate. Here, we...
0
0
0
RT @EinsteinMed: .@NEWS12BX @LindsayTanney featured Einstein’s Medical Scientist Training Program (MSTP) in a new #RebuildingTheBronx segme….
0
5
0
Super excited about this!.
Interested in phages? Live in the Bay Area & feel like spending a morning at Stanford?. You're invited to a special phage seminar with 2 of my favorite phage profs, visiting us all the way from Europe: . Speakers: Joana Azeredo (University of Minho) @azeredo_joana & Martin J.
0
0
1
RT @colinhillucc: Really love this story from the lab that shows that Faecalibacterium prausnitzii can package its own genome in bite-size….
0
6
0
Go @flamholz!.
A warm welcome to Rockefeller, Avi!. Avi Flamholz's lab will focus on the role of soil microbes in the carbon cycle, with the goal of unlocking new ways to harness the power of microbes to slow down or reverse rising global temperatures.
0
0
1
RT @azeredo_joana: 📢 Historic day for Portugal and phage therapy! 🇵🇹 A framework was approved, allowing magistral preparation of personali….
0
60
0
Tremendous advance for sequence analysis. Will be integrating to all workflows. Go @thesteinegger @milot_mirdita.
This is a foundational achievement. MMseqs2-GPU virtually *erases* the bottleneck of homology search in many workflows, including structure prediction. @sacdallago @milot_mirdita @thesteinegger, Felix, Bertil, and Alejandro too!.
0
0
3
RT @AsafLevyHUJI: I am excited to share our work "Systematic discovery of antibacterial and antifungal bacterial toxins" that is published….
0
91
0
Our AI for viral annotation review is out! I spent much of my dissertation discussing the opportunities and challenges that new AI methods pose for viral annotation. It is an exciting time!! @microbegrrl @mbiojournal @ASMicrobiology.
Improving viral annotation with artificial intelligence. 1/ This review highlights the potential of using artificial intelligence, specifically protein language models (pLMs), to tackle one of virology's toughest challenges: the annotation of viral sequences that lack homology
0
3
5
RT @steve_biller: Two research technician positions are now available in my lab at Wellesley College - come help us learn more about microb….
0
24
0
Very exciting work form the @tatta_bio team.
Introducing the 😱OMG dataset and 🤖gLM2! OMG is a massive Open MetaGenomic corpus totaling 3.1Tbp of data and 3.3B proteins. We train gLM2, the first mixed-modality genomic language model, on OMG. 1/🧵 #BioML #AI4Science.
0
0
0
Very excited to see more phage annotation tools leverage protein language models. Exciting stuff!.
It has been public for a while, but with #VoM2024 starting, today is as good as any to introduce our new tool phage genome annotation tool Phold. Phold uses structural homology to assign functions to phage proteins (1/n)
0
0
3
Awesome work and very excited to see the open-source. Can't wait to see how it does with phage proteins.
We have trained ESM3 and we're excited to introduce EvolutionaryScale. ESM3 is a generative language model for programming biology. In experiments, we found ESM3 can simulate 500M years of evolution to generate new fluorescent proteins. Read more:
0
0
0
RT @RajeevAlur: We are looking for postdocs to collaborate with Penn Medicine faculty for an exciting new project on Trustworthy AI for cli….
0
12
0
#phage remember this cover.
Jump into our June issue. 🧊 Carbon degradation in thawing peatlands.🌋 Carbon fixation in hydrothermal vents.🧪 Personalized phage therapy.🛑 Prevention of future mpox outbreaks.👄 Oral bacteria and health outcomes. and more, here 👇.
0
0
0
RT @NatureMicrobiol: Our June issue has gone live. In this month's editorial, we advocate for phage therapy as a strategy to combat the glo….
0
81
0