Yuri Pritykin
@YuriPritykin
Followers
874
Following
950
Media
32
Statuses
538
Assistant Professor @Princeton Lewis-Sigler Institute for Integrative Genomics (https://t.co/KhwYMNwZKd) and @PrincetonCS
Joined April 2014
Comp bio postdoc positions available! Come work with us @Princeton on computational method development and applications in regulatory genomics, genome editing, immunology and cancer, in mouse and human: https://t.co/4RNlQIQwxJ Please share among all those who may be interested!
pritykinlab.github.io
Pritykin Lab at the Lewis-Sigler Institute for Integrative Genomics and the Department of Computer Science at Princeton University.
1
11
26
Huge congrats to Gabe @gabriel_a_d_ on a fantastic PhD defense last week!!! The 1st PhD from the lab! So proud of his many research achievements e.g.: https://t.co/we4NhmpSZQ
https://t.co/TTDLiNHcrm
https://t.co/mQLFpJYZ4C He's bound to do amazing things ahead!
1
0
6
Continuing with single cells! Yuri Pritykin’s lecture, “Overview of Principles and Applications of Single-cell Genomics in Immunology” takes participants through available technologies and what they can tell us. #AAIcourses #ComputationalImmunology
0
1
4
Postdoctoral position in computational biology @YuriPritykin Princeton University See the full job description on jobRxiv: https://t.co/rXH5YrRSJD
#ScienceJobs
https://t.co/Ys2Jpojk6M
jobrxiv.org
Post a job in 3min, or find thousands of job offers like this one at jobRxiv!
0
1
3
Happy to present today at AAI Introduction to Computational Immunology Course today at UPenn
0
0
5
Excited to share our review on Thetis cells and dendritic cells, and their roles in mucosal tolerance and immunity @NatImmunol
https://t.co/hJwimUfExh
https://t.co/oI6d1GsgZz
4
40
134
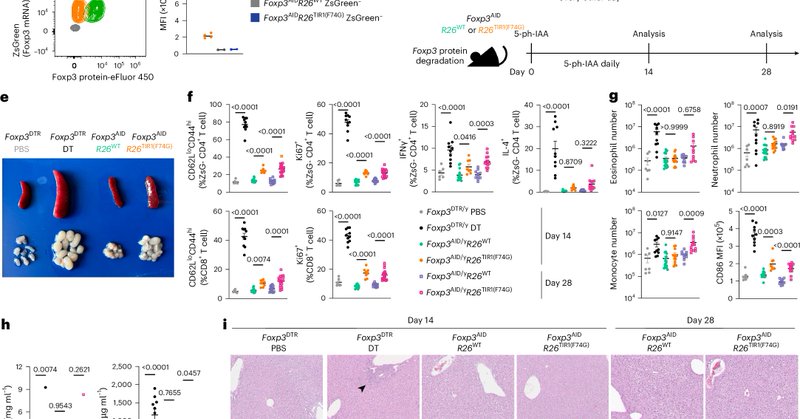
Temporal and context-dependent requirements for the transcription factor Foxp3 expression in regulatory T cells @NatImmunol @YuriPritykin Rudensky @Princeton @MSKCancerCenter
https://t.co/QDYZlL9bZu
0
16
85
Our paper is out today at @NatImmunol : https://t.co/lYTWW6s92m Very timely! We did extensive exploration of Foxp3 in Treg cells, using inducible protein degradation in vivo. Wonderful collaboration led by Sasha Rudensky and Wei Hu, computation led by talented @gabriel_a_d_
2
1
6
Extremely happy about the Nobel prize recognizing regulatory T cells! A fascinating concept with so many potential clinical applications. Very lucky to have been working on the genomics of Treg cells in collaboration with the giant in the field, Sasha Rudensky @MSKCancerCenter !
BREAKING NEWS The 2025 #NobelPrize in Physiology or Medicine has been awarded to Mary E. Brunkow, Fred Ramsdell and Shimon Sakaguchi “for their discoveries concerning peripheral immune tolerance.”
0
0
13
Excited to share our new study: Single-cell multiomics reveals archetypal regulatory programs shared across CD4 and CD8 T cell subsets in viral infection bioRxiv: https://t.co/ex73ak8RzL 🧵 Key findings below 👇
0
0
2
Excited to share our new study: Single-cell multiomics reveals archetypal regulatory programs shared across CD4 and CD8 T cell subsets in viral infection bioRxiv: https://t.co/ex73ak8RzL 🧵 Key findings below 👇
biorxiv.org
T cells protect against pathogens and tumors and can differentiate into various functionally distinct subsets. While each subset exhibits a characteristic epigenomic and transcriptional profile,...
Single-cell multiomics reveals archetypal regulatory programs shared across CD4 and CD8 T cell subsets in viral infection https://t.co/DyJImQYXdd
#biorxiv_immuno
1
4
12
Metadomain and metaloop genome interactions in mammalian T cells https://t.co/GPtPWPsU2o
#biorxiv_genomic
biorxiv.org
Recent studies have advanced our understanding of chromosomal organization and its principal role in gene regulation. However, most analyses have focused on short-range interactions (<2 Mb), limiting...
0
1
7
Excited for a major milestone in a collab effort led by @jengreitz to map enhancers & interpret variants in the human genome: The E2G Portal https://t.co/O9CRC1gRsf collates predictions of enhancer-gene regulatory interactions across >1,600 cell types & tissues. Use cases 👇1/
6
30
122
Postdoctoral position in computational biology @YuriPritykin Princeton University See the full job description on jobRxiv: https://t.co/rXH5YrRSJD
#ScienceJobs
https://t.co/6OsnWgzRd9
jobrxiv.org
Post a job in 3min, or find thousands of job offers like this one at jobRxiv!
0
1
3
This work is a wonderful collaboration led by amazing Sarah Walker, with Joris van der Veeken and Sasha Rudensky. More details in the preprint: https://t.co/ex73ak8RzL Looking forward to any feedback! 🙌
biorxiv.org
T cells protect against pathogens and tumors and can differentiate into various functionally distinct subsets. While each subset exhibits a characteristic epigenomic and transcriptional profile,...
1
0
1
In sum, we provide: – A resource and framework for high-resolution T cell regulatory analysis. – Archetypal regulatory decomposition of core T cell functions across lineages and infection contexts. – Insights into shared regulatory logic of CD8 progenitors and CD4 Tfh cells.
1
0
0
A key challenge is to decompose regulatory programs driving overlapping core T cell functions e.g. self-renewal, expansion, cytotoxicity, cytokine production. For this, we did archetypal analysis. It revealed that CD8 progenitors combine CD8 exhaustion & CD4 Tfh archetypes.
1
0
0
Surprisingly, these progenitor CD8 T cells were transcriptionally & epigenomically most similar to CD4 Tfh (follicular helper) cells. Despite distinct lineages, they shared activity of CXCR5, BCL6, TOX, TCF1, ID3 and PD-1. This suggests converging cellular phenotypes.
1
0
0
Notably, fundamentally and translationally important Tcf1⁺ progenitor CD8 T cells formed a shared population across acute & chronic infection. These results help reconcile recent studies of several related populations
1
0
0
We profiled 31k+ T cells with scATAC+RNA-seq during acute & chronic viral infection and built a 285-sample ATAC-seq atlas. This enabled robust identification of all major T cell states: naive, effector, memory, Treg, Tfh, exhausted & progenitor, shared across infection conditions
1
0
0