Xiang Zhou
@xiangzhoulab
Followers
653
Following
48
Media
0
Statuses
33
Genetics, Genomics, Statistics
Ann Arbor
Joined January 2011
RT @GenomeBiology: HIPPO, a heterogeneity-inspired preprocessing tool for analyzing RNA-seq data with UMIs, from Kim, @xzlab_org, and @Meng….
0
13
0
Very excited on this work with Mengjie Chen @MengjieChen6.
First day on Twitter! I want to share with everybody my recent work with my student Tae Kim and my long time collaborator Dr. Xiang Zhou @xzlab_org on single cell 10X UMI data.
1
0
7
RT @MengjieChen6: First day on Twitter! I want to share with everybody my recent work with my student Tae Kim and my long time collaborator….
biorxiv.org
Analysis of scRNA-seq data has been challenging particularly because of excessive zeros observed in UMI counts. Prevalent opinions are that many of the detected zeros are “drop-outs” that occur...
0
36
0
RT @GenomeBiology: A benchmarking of dimensionality reduction techniques for scRNA-seq data from @SunShiquan, @xzlab_org and co. They compa….
0
22
0
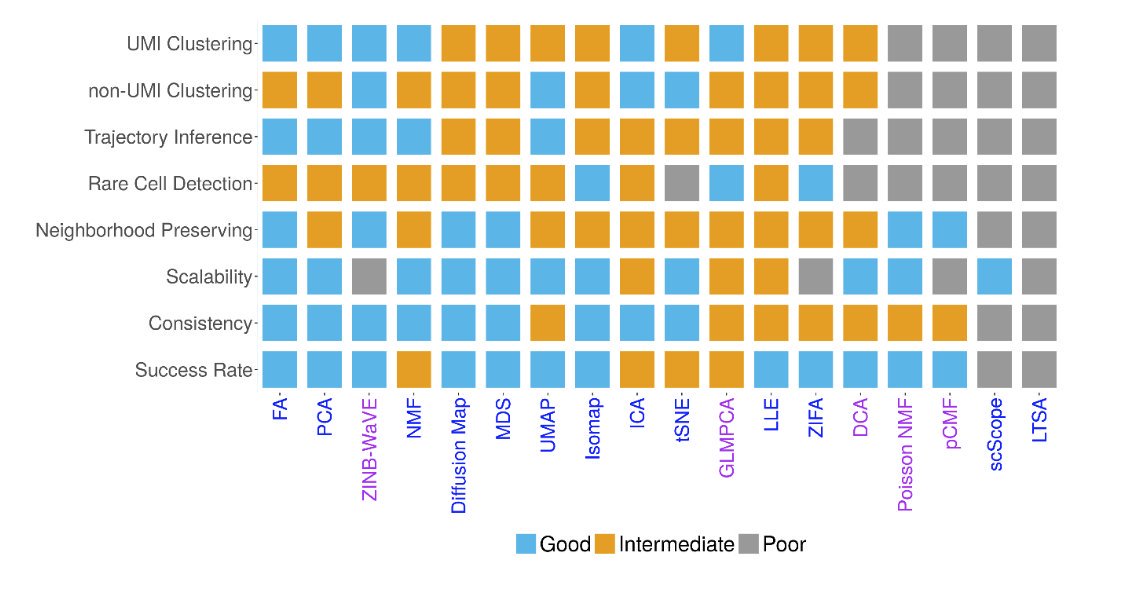
Exciting work lead by @SunShiquan; we were also quite surprised to see the results.
In a comparison of 18 dimension reduction/matrix factorization methods on 30 publicly available Single Cell RNAseq data sets, the simplest and oldest methods do surprisingly well compared to the new kids on the block. Nice work from Zhou lab @bioconductor
0
9
17
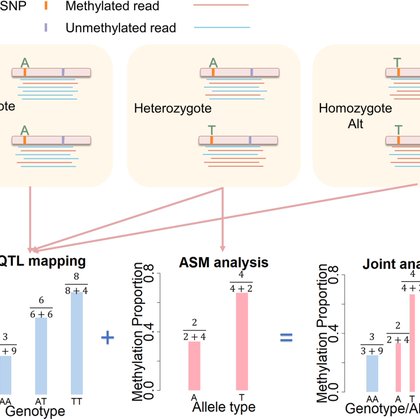
Exciting collaboration with @jtung5 on a computational method for integrative allelic specific methylation analysis and mQTL mapping, with applications to baboons and wolves
genomebiology.biomedcentral.com
Identifying genetic variants that are associated with methylation variation—an analysis commonly referred to as methylation quantitative trait locus (mQTL) mapping—is important for understanding the...
1
3
21
RT @jtung5: We're hiring! Looking for a lab tech to join us in beautiful Durham, N.C. to move forward our work on genomics and behavior in….
0
50
0
RT @LB_Barreiro: Excited to share our most recent work led by @GeneScientist17: Natural selection contributed to immunological differences….
0
43
0
RT @BrandonL_Pierce: Postdoc position available in my group at Univ of Chicago. Work at the intersection of genetic, molecular, & environ….
0
32
0
RT @NatureGenet: Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions (Urbut et al.)….
0
85
0
Our latest with Mengjie Chen on imputing single cell RNAseq data while preserving single cell level expression variability: @GenomeBiology.
0
17
31
RT @lorin_crawford: Our paper on the MArginal ePIstasis Test. Thanks for helping to get this out, @PLOSGenetics!
journals.plos.org
Author summary Epistasis is an important genetic component that underlies phenotypic variation and is also a key mechanism that accounts for missing heritability. Identifying epistatic interactions...
0
7
0
RT @marchini: The Oxford Statistics Phasing Server will phase your high-coverage human sequence data using the HRC panel .
0
25
0
RT @jtung5: Really nice coverage of our recent work on noninvasive genome resequencing by the talented Cristy Gelling! .
0
9
0
RT @blekhman: Please RT: Postdoc position available in my lab! Work at the interface of human genomics & microbiome. Email/DM me! https://t….
blekhmanlab.org
0
88
0
RT @WenXqwen: Our latest w/ @rogerpique @fluca2406 on integrative genetic analysis Find some cool nuggets in data….
0
10
0
RT @grg_perry: An evolutionary medicine perspective on Neandertal extinction. Our new #bioRxiv preprint! https://t.….
0
30
0
RT @biorxivpreprint: Normalization of Single Cell RNA Sequencing Data Using both Control and Target Genes #bioRxiv.
0
9
0