Ben WoOdcroft
@wwood
Followers
453
Following
202
Media
14
Statuses
199
Yet another microbial bioinformatician, group leader, ARC Future Fellow https://t.co/n4uQkFfRWL
CMR, QUT, Brisbane
Joined September 2008
Many many to thank, particularly @aroney_samuel,.@RossenZhao, Joshua Mitchell, Rizky Nurdiansyah Mitchell Cunningham @CoralSymbioses Gene Tyson @CMR_QUT and dozens of people who have helped with the software, ms, and everyone who tolerated my enthusiasm.
1
0
1
A new @rust4bio approach also helps - conserved regions are already aligned to each other so distance calcs become a vector similarity search problem. Big distance => novel species. Props to for awesome PR.
github.com
Bioinformatician at RasmussenLab, University of Copenhagen - jakobnissen
1
0
0
Fast and RAM-efficient since most raw reads are swiftly ignored. We optimise an up-front DIAMOND BLASTX-based method. Thanks @bbuchfink / Serratus for makeidx
1
0
0
Out in @NatureBiotech: Metagenome taxonomy profilers usually ignore unknown species. SingleM is an accurate profiler which doesn't, even detecting phyla with no MAGs. Profiles of 700,000 metagenomes at A 🧵
1
23
43
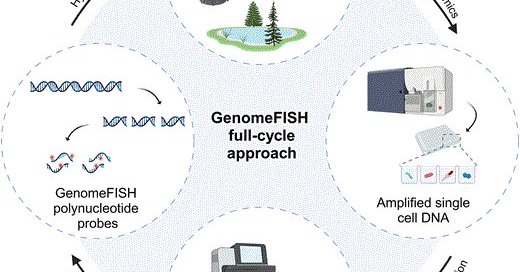
RT @Pam_Engelberts: Very excited to share the first paper out of my Postdoc @CMR_QUT:. GenomeFISH: genome-based fluorescence in situ hybrid….
academic.oup.com
Abstract. Fluorescence in situ hybridization (FISH) is a powerful tool for visualizing the spatial organization of microbial communities. However, traditio
0
4
0
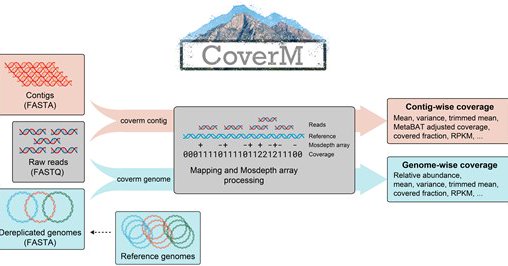
RT @CMR_QUT: Our newest tool, CoverM, a unified software package which calculates several coverage statistics for contigs and genomes in an….
academic.oup.com
AbstractSummary. Genome-centric analysis of metagenomic samples is a powerful method for understanding the function of microbial communities. Calculating r
0
8
0
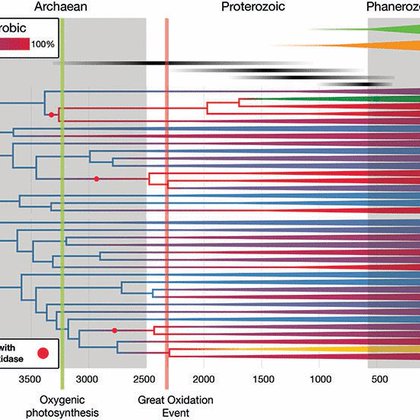
RT @ConversationEDU: Until now, it's been very hard for scientists to establish a detailed timeline of the early evolution of bacteria. @ww….
theconversation.com
Until now, it’s been very hard for scientists to establish a detailed timeline of the early evolution of bacteria.
0
1
0
We used Great Oxidation Event as a planet-sized "fossil" to add ancient dates to the Bacterial tree of life. The Conversation and @ScienceMagazine articles show oxygen was used by non-cyanos before that cataclysm, surprisingly.
science.org
Microbial life has dominated Earth’s history but left a sparse fossil record, greatly hindering our understanding of evolution in deep time. However, bacterial metabolism has left signatures in the...
1
2
1