Winnie Tan
@winn5tan
Followers
252
Following
722
Media
37
Statuses
398
🔬 Postdoc w/ @ShabihShakeel @WEHI_research🎓 PhD w/ @GenomeStability @SVIResearch👩🔬 Biochemist interested in cancer research 🇲🇾 🇦🇺views are my own 🧪🧬
Melbourne, Victoria
Joined November 2011
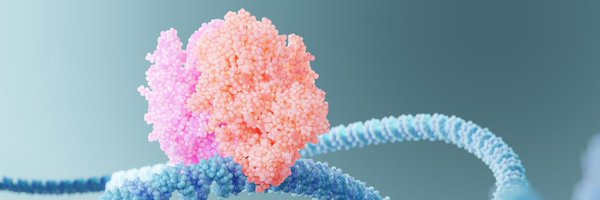
Excited to share our MORC2 paper out in @NatureComms! Feel free to read the open access version here — https://t.co/STblTanLSk
Excited to share the 1st preprint from @ShabihShakeel lab @WEHI_research on oncoprotein MORC2 that is now available on bioRxiv! Here’s a thread on our main findings on how MORC2 phosphorylation influences DNA compaction and its implications in cancer🔬📜
1
8
15
@nhmrc @ShabihShakeel @WEHI_research @CCeMMP @JeKyungRyu1 @HariVenugopal2 @MonashUni @jieqiong_lou @Liz_Hinde_ @UniMelb @DArcy_Lab_UTD @keeeeeeeeeeenan ..Alex, Tim, Andrew, Jumana, Vineet, Laura C, Laura D, Ching-Seng, @Vervoort_Lab, Gordon, @BlewittMarnie, @rhyscienceallan & @DavidovichLab. Thanks for everyone's help in the paper!
0
0
5
Massive thanks to funders @nhmrc & CMT Australia, @ShabihShakeel @WEHI_research @CCeMMP, @JeKyungRyu1, Jeongveen, Kyoung-Wook, JoonYoung (Seoul National Uni), @HariVenugopal2 @MonashUni, @jieqiong_lou, @Liz_Hinde_ @UniMelb, Prabavi, @DArcy_Lab_UTD, Tobi, @keeeeeeeeeeenan, Pedro..
1
0
4
Researchers learned a function of enzyme MORC2, that helps organise DNA and turn off genes when not needed. Mutations in MORC2 cause Charcot-Marie-Tooth disease, a rare condition that weakens muscles. These findings could lead to new treatments. https://t.co/oDzT0FB7rd 🧵1/2
1
3
11
🧬New from crystal structures from our lab in EMBO J! We reveal how the FANCM enzyme, an emerging therapeutic target it cancer, has evolved to specifically recognise branched DNA and activate the Fanconi anaemia pathway of DNA repair. https://t.co/tnVzM0R8RC 🧵👇
embopress.org
imageimageFANCM employs a sophisticated dual recognition mechanism in genome maintenance, using distinct DNA-binding domains to identify and move around on branched DNA structures. This study reveals...
1
7
23
Excited to share our pre-print on the molecular architecture of heterochromatin in human cells🧬🔬 w/ @jpkreysing, Johannes Betz,Turoňová lab, @MarinaLusic, Hummer lab, @Beck_Laboratory, @MPIbp 🔗 Preprint here: https://t.co/0tTxZcu0Vz
5
120
414
Big announcement🎙️Registration to our @arc_gov_au CCeMMP Research Symposium 2024 is now open!🥳 The symposium will be held on 11-12th Nov, in Melbourne 🇦🇺. We invite the scientific community to attend and participate!🔬 More details & register here: https://t.co/STw5pQf2x7
0
12
21
11/ Thanks to JeongVeen, @HariVenugopal2, JieQiong, Prabavi, Pedro, Toby, Kyuong-Wook, @keeeeeeeeeeenan, Alexandra, Andrew, Jumana, Vineet, Laura D, Ching-Seng, Laura C, @DavidovichLab, @Vervoort_Lab, Gordon, @BlewittMarnie, @rhyscienceallan, @Liz_Hinde_, Sheena and @JeKyungRyu1
0
0
5
10/ A big thank you to our amazing team and collaborators for their hard work and dedication to this project!👏#epigenetics #biology Our findings were also reported by a similar preprint👇, glad to see that we achieved similar findings at the same time! ☺️ https://t.co/GehhEmcEaX
biorxiv.org
The eukaryotic microrchidia (MORC) protein family are DNA gyrase, Hsp90, histidine kinase, MutL (GHKL)-type ATPases involved in gene expression regulation and chromatin compaction. The molecular...
1
0
2
9/ The findings from our study emphasize the importance of MORC2 phosphorylation in regulating chromatin dynamics and its potential as a cancer therapeutic target. We’re excited about the possibilities this opens up for future cancer treatments 👩🔬👨🔬💊🏥
1
0
0
8/ These findings support a new model for chromatin remodelling by MORC2, wherein low-affinity DNA binding sites on the CTD facilitate sequence-independent genome surveillance. Upon a compaction signal, high-affinity sites trap the genome region, forming a DNA loop ✨✨
1
0
0
7/ Finally, we use atomic force microscopy to visualise the MORC2-DNA complex, and found compaction of DNA mediated by the ATP hydrolysis, and modulated by CTD phosphorylation 💥🧬🔬
1
0
2
6/So why is MORC2 clamping DNA? To answer that q, we use single molecule fluorescent microscopy, which allows us to visual DNA sliding. And boom! 💥We discovered a DNA compaction activity of MORC2, where it forms compacted DNA 💥🧬
1
0
3
5/ To assess if CTD domain of MORC2 is required for DNA binding, we perform competition DNA assays with linear and circular DNA. Surprisingly, circular DNA helps it clamp tighter to DNA! This suggest a potential role for MORC2 as a sliding DNA clamp, which required its CTD 🛝🧬
1
0
1
4/ Combining cryo-electron microscopy, crosslinking and HDX mass spec, we characterise MORC2 binding to DNA and show that there are several conformational changes across CC1, CW and CC4 domains, hinting multiple DNA binding at different sites 🧬
1
0
1
3/ Because MORC2 phosphorylation was associated with gastric cancer, we purified full-length MORC2 and assessed its phos state. We identified a phosphate interacting motif within the C-terminal domain (CTD) of MORC2, whose phosphorylation state modulates ATP hydrolysis activity
1
0
1
2/ We ask where does MORC2 bind to across the genome? Using ChIP-seq, ATAC-seq and FLIM-FRET, we found that MORC2 peaks are most abundant at the promoter region, suggesting its binding to open chromatin region associated with DNA, as opposed to H3K9me3 associated closed chromatin
1
0
1
1/🧵MORC2, a chromatin remodeling ATPase, plays a crucial role in epigenetic regulation and DNA damage response. Variants in MORC2 can lead to defects, promoting cancer progression. We aimed to uncover the molecular mechanisms behind this at the cellular to atomic level #cryoEM🔬
1
0
1
Excited to share the 1st preprint from @ShabihShakeel lab @WEHI_research on oncoprotein MORC2 that is now available on bioRxiv! Here’s a thread on our main findings on how MORC2 phosphorylation influences DNA compaction and its implications in cancer🔬📜
biorxiv.org
Variants in the poorly characterised oncoprotein, MORC2, a chromatin remodelling ATPase, lead to defects in epigenetic regulation and DNA damage response. The C-terminal domain (CTD) of MORC2,...
1
9
31
@WEHI_research is hiring! Share with your colleagues! Laboratory Head - RNA Biology
nature.com
An excellent opportunity is available for a Laboratory Head to provide leadership and focus for RNA Biology at WEHI.
0
17
13