Soham Gadgil
@soham_gadgil
Followers
68
Following
209
Media
6
Statuses
28
Ph.D. student @uwcse, previously @Microsoft MSCS @Stanford BS in Computer Engineering @GeorgiaTech Working on explainable AI for healthcare.
Seattle, WA
Joined January 2016
Medical #AI errors “can directly impact people’s health and even determine life-altering outcomes.” In @natrevbioeng, @UW #UWAllen’s @ChanwooKim, @soham_gadgil & @suinleelab emphasized the importance of transparency in medical models. #ResearchMakesAmerica
washington.edu
In a recent paper, University of Washington researchers argue that a key standard for deploying medical AI is transparency — that is, using various methods to clarify how a medical AI system arrive...
0
4
9
Nothing more fun than working with brilliant students @uwcse @ChanwooKim_ & @soham_gadgil on our Nature Reviews bioengineering paper!🎉We review challenges & opportunities for making medical AI trustworthy through transparency in data, models & deployment. https://t.co/ZZBdtgixVz
0
5
16
It was a pleasure working on this review with amazing co-authors @ChanwooKim_ and @suinleelab from @uwcse! UW News: https://t.co/8hBNDcqodT (4/4)
washington.edu
In a recent paper, University of Washington researchers argue that a key standard for deploying medical AI is transparency — that is, using various methods to clarify how a medical AI system arrive...
0
0
3
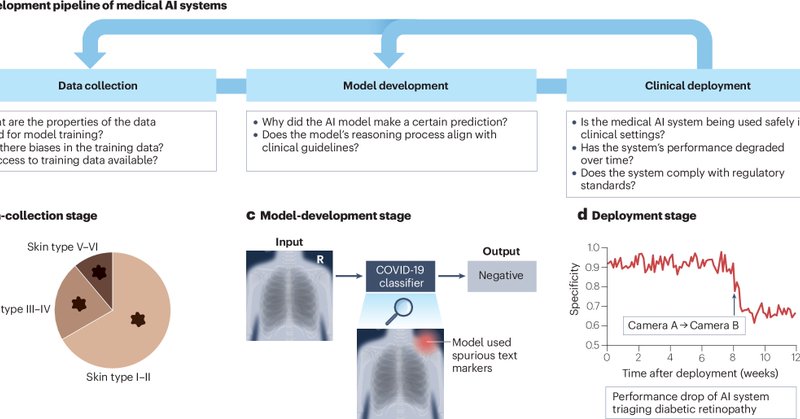
In this review, we examine the latest advancements in transparency for medical AI across the entire pipeline, from the training data to the model development and real-world deployment. We also provide insights to ensure transparency in emerging AI models such as LLMs. (3/n)
1
0
0
As AI systems become more widespread, it is essential to ensure they remain reliable, trustworthy, and explainable, especially in high-stakes domains such as healthcare. We argue that transparency should be a crucial component of medical AI development and deployment. (2/n)
1
0
0
Excited to share that our review paper on "Transparency of medical artificial intelligence systems" has been published in Nature Reviews Bioengineering! Paper: https://t.co/QThjxdH8uK (1/n)
nature.com
Nature Reviews Bioengineering - Artificial intelligence (AI) models are being applied more often across a range of biomedical domains to support clinical decision-making and therapeutic strategies....
1
0
1
🚀 We’re hiring! Multiple postdocs & a program manager to push the frontiers of explainable AI in cutting-edge biomedical research—Alzheimer’s, aging, cancer & medical AI. Start immediately. Friends, please RT🙏 Learn more: https://t.co/pAR8O6EKNP
#postdocjobs #AI #Biomedicine
0
15
35
Congrats to @soham_gadgil on this awesome pre-print — not only does this method for vision interp use SotA techniques, but really importantly (I think) emphasizes the human-in-the-loop. So much of the value of interpretability is as a lens for looking carefully at your data
1
1
4
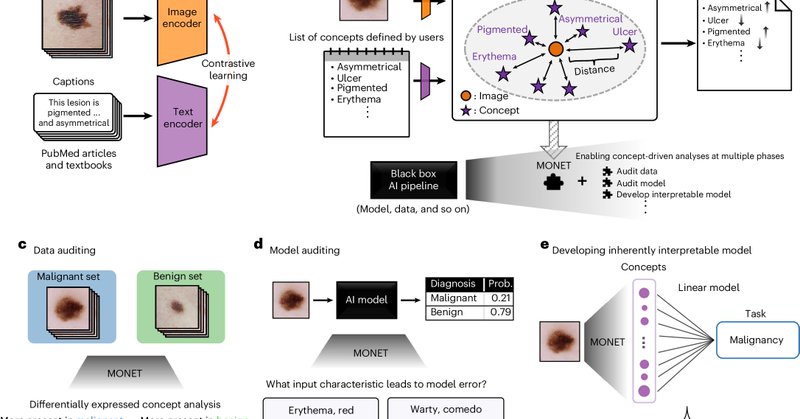
Concept is all you need! @uwcse features our @NatureMedicine paper on extracting concepts from dermatology images via foundation models, with brilliant first author @ChanwooKim_ and exceptional co-senior author @RoxanaDaneshjou at @StanfordDBDS. Details:
news.cs.washington.edu
In a recent paper published in the journal Nature Medicine, a team of researchers at the University of Washington and Stanford University co-led by Allen School professor Su-In Lee introduced a...
1
7
40
It was a pleasure working with co-first author Alex DeGrave along with advisors @suinleelab and @RoxanaDaneshjou! Check out our paper at:
medrxiv.org
Recent advances in Artificial Intelligence (AI) have started disrupting the healthcare industry, especially medical imaging, and AI devices are increasingly being deployed into clinical practice....
0
2
2
Excited to share that our paper "Discovering mechanisms underlying AI prediction of protected attributes via data auditing" won the best paper runner up award at the Data Curation and Augmentation in Medical Imaging Workshop at CVPR 2024! (1/2)
2
0
8
Excited to announce the inaugural AIMBA (AI Meets Biology of Aging) 2024 meeting on May 22, 2023, 9-2pm PT, organized by @UW @NathanShockCtrs ! Featuring four keynote speakers: Profs @mariabrbic, Anne Brunet, Vadim Gladyshev, @james_y_zou. Check it out! https://t.co/O8eWzYTleZ
1
15
43
Excited to see our @NatureMedicine paper out today led by @ChanwooKim_ with my co-senior author @suinleelab and an amazing team! We used a dermatology foundation model to enable explainable and transparent AI - from auditing datasets to models.
nature.com
Nature Medicine - By learning to pair dermatological images and related concepts in a self-supervised manner, a visual-language foundation model is shown to have comparable performance to...
6
33
176
Happy to share that our paper on leveraging foundation models to foster the explainability and transparency of medical AI has been published today in @NatureMedicine! Check it out here:
nature.com
Nature Medicine - By learning to pair dermatological images and related concepts in a self-supervised manner, a visual-language foundation model is shown to have comparable performance to...
1
15
48
Happy to share that DIME has been accepted at ICLR 2024! This work was in collaboration with the amazing co-author @ianccovert and advisor @suinleelab #ICLR2024
How to perform dynamic feature selection without assumptions about the data distribution or fitting generative models? We develop a learning approach to estimate the conditional mutual information in a discriminative fashion for selecting features. https://t.co/6dCHlJJA9m
0
1
20
It was a pleasure working on this project with co-first author @ianccovert and advisor @suinleelab from @uwcse! (7/7)
0
0
0
We generalize our approach to incorporate prior information beyond the main features. Here, we again prove that our procedure recovers a modified version of the CMI at optimality and show that this modification helps in improving performance. (6/n)
1
0
0
DIME is agnostic to the model used for training the network and we investigate the role of modern architectures in improving performance in DFS. Specifically, we find that for image data, our method benefits from using ViTs rather than standard CNNs (5/n)
1
0
0
Taking inspiration from adaptive submodular optimization, we also show how to adapt our CMI-based approach to scenarios with non-uniform feature costs while retaining high performance. (4/n)
1
0
0
Comparing to multiple baselines (static/dynamic feature selection methods), we find that our method (DIME) provides consistent gains across a variety of image and tabular datasets. We include real-world medical datasets where this approach is practically useful. (3/n)
1
0
0