Ronglai Shen
@ronglais
Followers
128
Following
233
Media
2
Statuses
42
Joined February 2009
🧪 Have tested on ~100 whole-slide images (~40M cells). 🔄 Actively developed — we're now optimizing parallel processing across hundreds of slides. 🙏 Huge thanks to the amazing team & all collaborators @ronglais @jsmithymd @KPanageas @MSKBiostats @TAMUstats 6/6
1
1
2
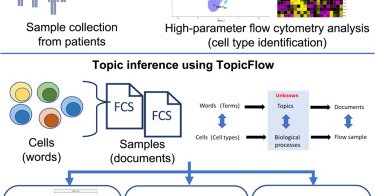
"Excited to share our paper, now online in @CellRepMethods. We analyzed 17M T cells from 138 PBMC samples from melanoma patients on nivo-ipi. https://t.co/gX81Krrmyk
#SingleCell #CancerImmunotherapy #flowcytometry
cell.com
Peng et al. introduce a computational framework, TopicFlow, to explore T cell population dynamics in peripheral blood samples obtained from patients with cancer receiving immune checkpoint inhibitor...
2
3
19
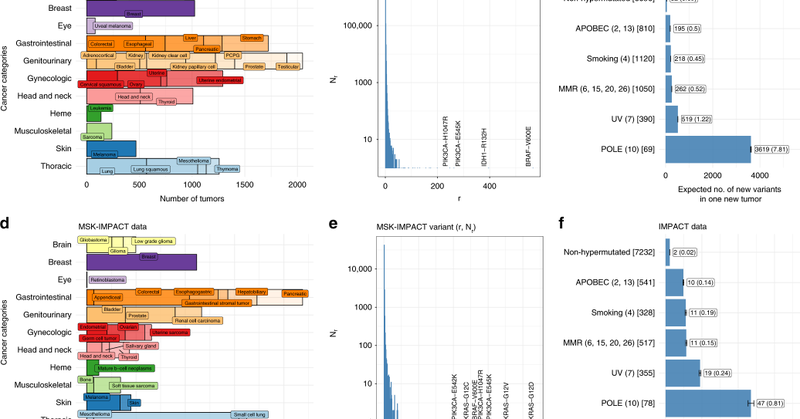
Using #genomic and #epigenomic meta-features to predict the tissue of origin of cancer from the hidden genome of #RareVariants: new #CancerResearch paper by @SaptarshiCh @CLaunderer @ColinBBegg @ronglais
@NatResCancer
https://t.co/Um1TXHoUWk
0
6
10
25 min of smart and funny people talking about culture (including delicious food 🙂) #StopAsianHate #DailyShow
2
2
9
I am voting for a future that believes in science and immigrants.
1
0
14
Probability estimation for unseen variants is the key insight to permit identification of lineage-dependency, critical for classifying primary site of origin of a tumor and clonality testing.
0
0
1
Method involves adaptation of Good-Turing frequency estimation (used in cracking the Enigma code) to variant probability estimation in the context of extreme data sparsity.
1
0
2
On average, 2 out of 3 variants detected in a new tumor in the MSK-IMPACT cohort have not been seen in the TCGA data.
1
0
0
@ColinBBegg @c7rishi @arorarshi @sloan_kettering Vast majority of variants cataloged in large-scale sequencing studies are rare, with >90% being singletons.
1
0
0
Our work in Nature Communications on mining the hidden iceberg of rare and unseen variants in the cancer genome using ecology and computational linguistic tools: https://t.co/jho5MfOJHs
@ColinBBegg @c7rishi @arorarshi @sloan_kettering #MSKimpact #Genomics
nature.com
Nature Communications - Sequencing cancer genomes reveals low frequency novel somatic variants without known function. Here, the authors leverage statistical methodology from the fields of...
2
5
15
Introducing 📦panelmap to visualize grouped data and circomap for multiple cohorts in R! https://t.co/dmK45lJ7lh
#dataviz #rstats check out an example from mtcars!
0
7
9
.@arorarshi, @CLaunderer and I were proud to represent @sloan_kettering at the First International Symposium of Mathematical and Computational Oncology these past few days! #ISMCO2019 #rstats
1
4
18
Another packed house for today’s @MSKPathology Special Seminar (“How Pathologists Can Learn to Stop Worrying and Love Statistics”) with @sloan_kettering’s Dr. Mithat Gönen #MSKPathology #MSKPathologyGrandRounds
1
4
25
THREAD: (1/10) Our work in JCO PO looked at NGS data from patients with met lung adeno. Some goals: (1) explore notion of molecular staging (within stage IV) using somatic mut & (2) see what drives poorer outcomes under current standard of care https://t.co/0zMtMTceEV
#MSKIMPACT
4
32
75
Our paper on genomic risk stratification in metastatic lung adenocarcinomas is now online https://t.co/Sx181LgcMe
@RielyMD @DSolit @MLadanyi
ascopubs.org
PURPOSEBroad-panel sequencing of tumors facilitates routine care of people with cancer as well as clinical trial matching for novel genome-directed therapies. We sought to extend the use of broad-p...
2
20
34
New manuscript with recs on how to normalize scRNA-Seq data. Main message: don't use log(CPM+1) transformation, it magnifies unwanted source of variability. For example, see tSNE plots of technical replicates below. For more see https://t.co/dGWFpN8Byq and thread by @sandakano.
4
146
309
Just donated to Epi-BioCyclers for @Cycle4Survival. So proud to be part of the community.
0
0
4