Radoslav Aleksandrov
@radaleksandrov
Followers
102
Following
788
Media
8
Statuses
39
Beaming cells to see what happens at @LGS_IMB #DNArepair #DNAreplication #Livecellimaging #Cancer #Anticancerdrugs
Joined March 2018
Easily one of the most beautiful live-cell imaging papers out there! Amazing contribution to chromosome tracking during mitosis with huge prospects for further discoveries concerning chromosome behavior. Check it out @NatureCellBio!
Have you ever wanted to track individual #chromosomes during #mitosis? To do live cell #karyotyping and map the inter- and intra-chromosomal contacts dynamics? Our recent work made that possible. Check out our @NatureCellBio paper: https://t.co/w6iaGC8a6n
0
0
0
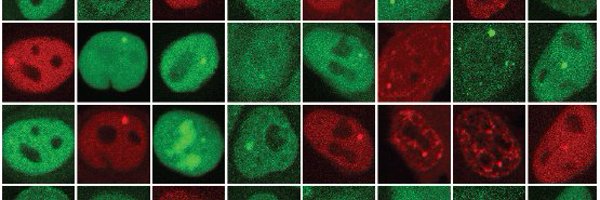
Contributing to efforts toward elucidating the response to replication stress, we provide a precise timescale of fork stalling and restart in live cells, at 30-s resolution (airyscan teaser below). https://t.co/cvLW0YaI5O
#replicationstress #DNArepair #ATR #replication #DNA #RPA
0
13
12
We are thrilled to present you our work on BRCA1-BARD1 @Nature showing that the complex has a direct role in long-range DNA end resection. With @CeppiIlaria @DelloStritto_MR and our collaborators @PabloHuertasLab @SylvieNoorderm1 and Seidel labs. https://t.co/xh3J7mtwaV
19
68
266
🥳Our research on the temperature sensitivity of bat antibodies has now been published! #bats #immunity #metabolism A lovely collaboration with Dr JD Dimitrov's group at @CRCordeliers, @nmnhs, @lab_jouvenet, @CyrilPlanchais 🦇 https://t.co/Tn8BiJGE5M
2
19
64
We welcome Bulgaria as EMBL’s newest prospect member state! This is an important milestone that opens up new opportunities for mutual exchange and collaboration. https://t.co/UqN00b1uU1
0
19
82
19/ PPS. We also invite those who seek further details regarding the mathematical modeling employed herein to check out: https://t.co/FW1cJW50C2
0
0
3
18/ PS. For those who want to brush up on their PARP1/PARPi knowledge (past & present), we invite you to read our recently published review on the matter: https://t.co/UthPqLBTk6
1
0
4
17/ Lastly, a HUGE THANK YOU AND CHEERS to all the amazing people involved in this work @petkanev @SylviaMarinova1 Irina Georgieva @MariaLukarska Dilyana Kirova @GeorgiDanovski @StoynoS @LGS_IMB and the BEST spinning disk @AndorTechnology
1
0
5
16/ All imaging data (terabytes and terabytes…) were analyzed and modeled in CellTool – a superb open-source software tailored to the analysis of live-cell imaging and micro-irradiation data created by @GeorgiDanovski #CellTool
https://t.co/RBWlw7XUkm
1
0
3
15/ … we are confident that such an approach may prove very useful in elucidating the properties and mechanisms of many other DNA repair-targeting drugs with the ultimate goal to deliver more effective and salutary therapies for patients.
1
0
3
14/ We believe that the approach outlined here could be used to interrogate the properties of PARP1 (and why not other proteins?) in various genetic backgrounds or in diverse model systems to tackle the ever-pressing issue of PARPi resistance. Further…
1
0
3
13/ …the PARP1 Retention Coefficient (PRC), which is a complex but precisely measurable characteristic of PARP1, could prove to be a useful predictor of PARPi efficacy in preclinical and clinical settings.
1
0
3
12/ Finally … and very importantly, we show that the extent of gross PARP1 retention at damaged chromatin sites (PRC) strongly corresponds to the delay in downstream repair events and PARP1 cytotoxicity in long-term clonogenic assays. Hence…
1
0
3
11/ A working model must not only explain a set of experimental results, but also harbor significant predictive powers. Using the strong correlation between PRC and PTC values upon complete inhibition, we managed to accurately predict the properties of hitherto unexplored PARPi.
1
0
3
10/ We then calculated the in vivo IC50 values (50% catalytic inhibition) for all evaluated PARPis.
1
0
3
9/ Finally, the PARP1 Inhibition Coefficient (PIC) quantifies the extent of PARP1 catalytic inhibition upon PARPi treatment. We calculate the PIC based on the amounts of ARH3, a key PAR-degrading enzyme, recruited to DNA damage sites.
1
0
3
8/ The PARP1 Trapping Coefficient (PTC) quantifies alterations in PARP1 exchange rates (turnover) following photobleaching (FRAP) of IR-induced DNA damage sites. We calculate the PTC as the halftime of fluorescence recovery in PARPi-treated cells vs. that in non-treated cells.
1
0
3
7/ The PARP1 Retention Coefficient (PRC) takes into account the gross kinetics of fluorescently-labeled PARP1 at IR-induced damage sites in living cells. We calculate the PRC as the ratio between the halftime of PARP1 removal in PARPi-treated vs. non-treated cells.
1
0
3
6/ To unravel this mechanism, we introduce a set of three straightforward and unbiased parameters which precisely quantify the alterations in PARP1 dynamics and activity in PARPi-treated live cells.
1
0
3
5/ Thus, PARPi cause PARP1 to unproductively bind and dissociate from DNA lesions multiple times, spending more time in its bound state due to allosteric trapping, which greatly increases overall PARP1 chromatin retention.
1
0
3