Helena Schulz-Mirbach
@mirbachh
Followers
237
Following
287
Media
18
Statuses
162
Metabolic engineer for a better future 👩🏼🔬. Former Bar-Even member and PhD student @erblabs 🥳🪄.
Joined December 2021
RT @PabloINik: #Synthetic C1 #metabolism in Pseudomonas putida enables strict formatotrophy and methylotrophy via the reductive glycine pat….
0
5
0
4) Many thanks to @BDronsella and @erblabs for all the interesting discussions. In addition, want to thank @haihe10, María Suárez Diez, and Elad Noor for constructive feedback on this work!.
0
0
1
3) My hope is that our work can serve as #starting point to compile selection strains into a readily accessible #collection. Let me know if you’d find that interesting!.
0
0
1
2) To make the metabolism schemes broadly accessible, you can download the .ai files from ! Can only recommend laminating those to sketch on when discussing anything metabolism :) .
github.com
Figures visualizing the central metabolism of E. coli - helenaschulzmirbach/ecoli-metabolism
0
1
4
RT @PabloINik: Implementing synthetic serine cycles in #Pseudomonas putida unlock methanol assimilation in our favorite bug with carbon con….
0
8
0
RT @mpi_marburg: 📢We are thrilled that our Microbes for Climate (M4C) Cluster of Excellence has been selected for the 2025 Excellence Strat….
0
27
0
RT @fidelormz: I'm truly excited to announce our new publication in @Nature unraveling a central picture of the Methyl-coenzyme M reductase….
0
39
0
RT @AriSatanowski: Excited to share a main project from my PhD, out now in @NatureComms! 📝.We've designed and brought to life the “CORE cyc….
0
6
0
RT @PabloINik: Seven Critical Challenges in Synthetic One-Carbon Assimilation and Their Potential Solutions—led by @FavoinoGiusi & @OscarPu….
0
11
0
RT @marchal_dg: 🌿🔬How can we fix #CO2 more efficiently in biological systems? Our new study introduces the CORE cycle, a synthetic pathwa….
nature.com
Nature Communications - Available enzymatic CO2 reduction strategies are not suitable for aerobic microorganisms and many industrial settings. Here, the authors design a new metabolic pathway that...
0
9
0
RT @sgqcheng: 📑📑📑#synthetic_biology @enricooorsi @mirbachh @PabloINik @erblabs present a workflow for designing versatile auxotrophic metab….
nature.com
Nature Communications - Auxotrophic metabolic sensors (AMS) are vital for bioengineering but are often time-consuming to develop. Here, the authors present a workflow for designing versatile AMS,...
0
2
0
RT @BDronsella: Check out this amazing work on engineering, characterizing and using six novel glyoxylate sensor strains led by @enricoorsi….
0
1
0
RT @PabloINik: Computation-aided designs enable developing auxotrophic #metabolic #sensors for wide-range glyoxylate and glycolate detectio….
nature.com
Nature Communications - Auxotrophic metabolic sensors (AMS) are vital for bioengineering but are often time-consuming to develop. Here, the authors present a workflow for designing versatile AMS,...
0
8
0
@NatureComms 9) It was a true pleasure to work with @enricoorsi , @haihe10 , @AriSatanowski , @PabloINik @erblabs and all co-authors involved. The origins of the strains go way back to @WeTheBarEvenLab and Charles Cotton creating the first strains - I am more than grateful to see it out !.
0
0
3
@NatureComms 8) The sensor strains are supposed to become a #community resource – so please feel free to reach out to us if you could apply them in your projects! This work would not have been possible without @enricoorsi doing a tremendous job at leading it.
0
0
2
@NatureComms 7) Of course, you can also use them to detect glyoxylate or glycolate in #environmental applications. For example, we could quantify how much glycolate was produced by #cyanobacteria during #photorespiratory growth based on the OD our sensors reached.
0
0
2
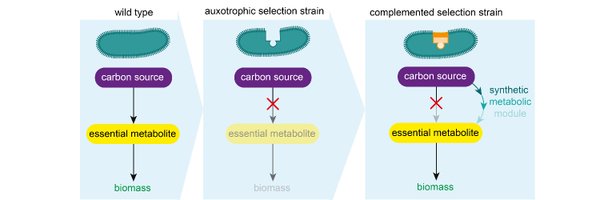
@NatureComms 6) What can you use those sensors for ? Amongst other things, to select for synthetic metabolic modules. As an example in the context of #C1assimilation, we engineered a module of the HOPAC cycle and show that we can determine the best #expression strength for the catalysts.
0
0
2
@NatureComms 5) Next, we characterised the strains by verifying expected metabolic fluxes by isotope tracing and determining their selective demands with #glyoxylate and #glycolate.
0
0
2