Ralph Mazitschek
@mazit
Followers
287
Following
650
Media
25
Statuses
212
ChemicalBiologists, all things small molecule at the corner of science and nature
Boston, MA
Joined March 2009
In the recent issue of @nchembio we report the development of #CoraFluors, a #novel class of bright and stable TR-FRET donors optimized for biological #application, including measuring the binding of drug molecules to their targets, directly in #livecells and in real time 🧪(1/n).
The cover of our November issue ( features a class of luminescent terbium-based probes called CoraFluors ( used for detecting ligand-bound protein targets by TR-FRET.
4
8
55
RT @CellChemBiol: Voices: @antolin_aa, Yimon Aye, @LironBarPeled, @Elena_De_Vita, @nata_dudkina, @MichaelCJewett, @hannahjanekc, @mazit, an….
0
7
0
RT @vicmarando: Excited to see this out! Congrats to @HammelForrest and the whole team!! @Chemist_Payne @mazit @HarvardMicro .Identificati….
pubs.acs.org
O-GlcNAc transferase (OGT) is an essential mammalian enzyme that binds thousands of different proteins, including substrates that it glycosylates and nonsubstrate interactors that regulate its...
0
9
0
RT @brian_b_liau: Excited to share 2 preprints, together showing how mechanisms of a molecular glue & E3 ligase cancer mutations serendipit….
biorxiv.org
Cancer mutations can create neomorphic protein-protein interactions to drive aberrant function[1][1]. As a substrate receptor of the CULLIN3-RBX1 E3 ubiquitin ligase complex, KBTBD4 is recurrently...
0
34
0
Rockstar work championed by @Chemist_Payne and Saki Ichikawa synergistically combines #CoraFluors and #Cyclimids into a powerful platform approach to accelerate degrader development and comprehensive characterization.
Our newest work in collaboration with Saki Ichikawa and @drcmwoo @mazit is now online @CellChemBiol! Here is a free-to-read link:.
0
4
20
Awesome collaboration with #BullockLab and @Chemist_Payne! Combining our complementary expertise, we shed new light on the CUL-3-KEAP1 complex, establish a straightforward assay to measure CUL3 binding to its adaptor proteins, and reveal CDDO’s activity as a “partial antagonist”.
The Bullock lab @CMDOxford has a new paper on KEAP1-CUL3 structure, TR-FRET and inhibition by CDDO. Great collaboration with Ralph Mazitschek @mazit DOI:
0
1
7
RT @DSzymborski: My Saturday fun project: using AI, every US president as a Pixar character.
0
13K
0
RT @Chemist_Payne: Preprint out! . KEAP1-CUL3 structure, an awesome new assay platform for BTB domain-containing proteins, and elucidating….
biorxiv.org
KEAP1 promotes the ubiquitin-dependent degradation of NRF2 by assembling into a CUL3-dependent ubiquitin ligase complex. Oxidative and electrophilic stress inhibit KEAP1 allowing NRF2 to accumulate...
0
4
0
RT @NatureComms: Authors @mazit @Mark_A_Tye @Chemist_Payne @dyann_wirth @LabOppermann @MGHCSB @HarvardChanSPH @broadinstitute @UniofOxford….
nature.com
Nature Communications - The development of antimalarials against the human liver and asexual blood stages is one of the top public health challenges. Here, the authors report a single-step...
0
7
0
P.S. Like many folks on #ScienceTwitter, we have also encountered hostile🗡️ referees during the review process of this manuscript. I am grateful🙏 to the editor for recognizing this and not taking the easy way out but recruiting additional referees. 12/.
0
0
6
Also thanks to our institutions and funding support @MGHCSB, @MGH_RI, @broadinstitute, @HarvardChanSPH, @broadinstitute, @UniofOxford, @HarvardCCB, @chemical_phd, @NIAIDFunding, @NSF, @scienceinboston 11/.
1
0
6
Nothing of this would have been possible without the awesome students and collaborators, @Mark_A_Tye, @Chemist_Payne, @kritika_singh24, @MariaMMota2, @ElizabethWinze1, @DerbyLabDuke, @dyann_wirth, @LabOppermann. 10/.
1
0
6
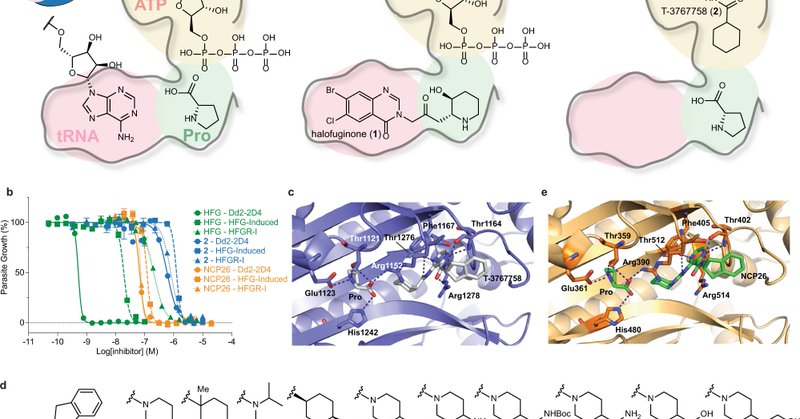
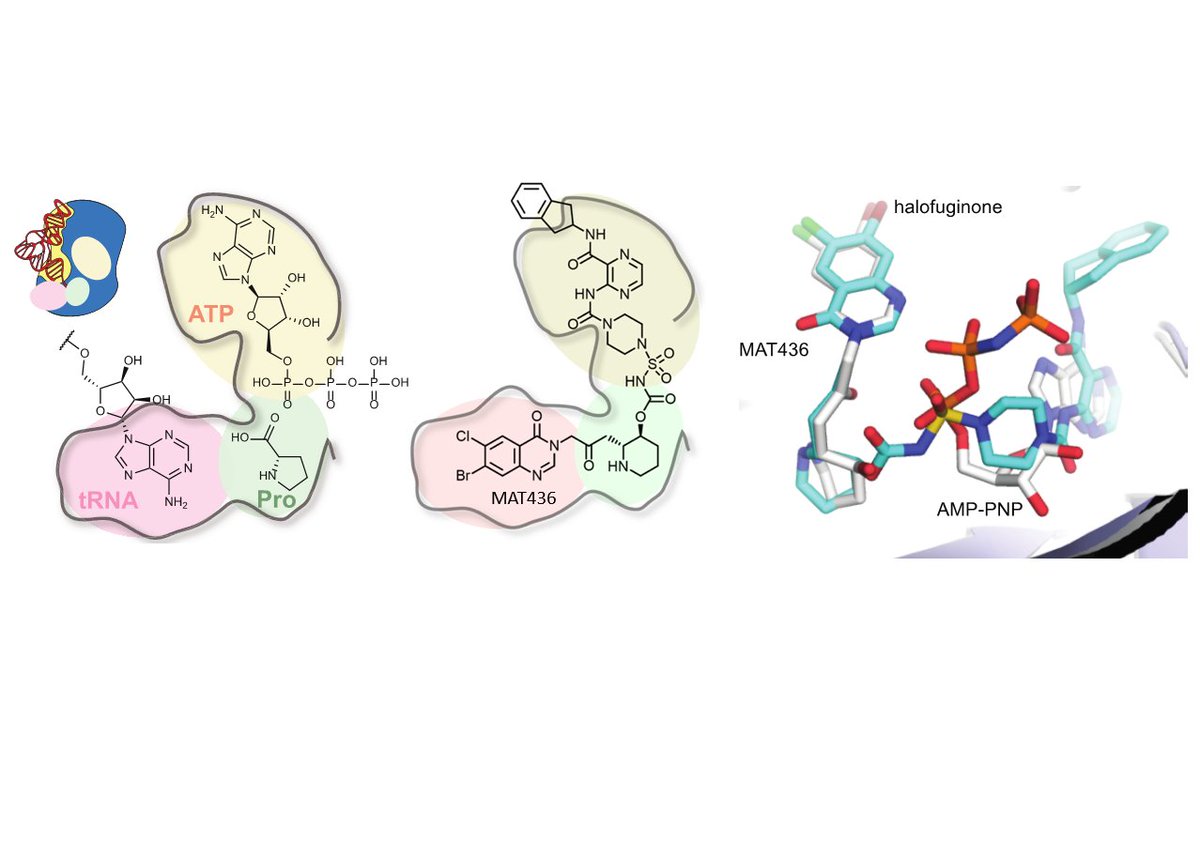
Using our new #CoraFluor TR-FRET ⚡️technology, we have developed a robust (Z’ > 0.9) mix-and-read🍸 ligand displacement assay for Plasmodium and human ProRS that is > 1000-fold more sensitive than current assays and enables facile determination of the mode of inhibition. 6/
1
0
6
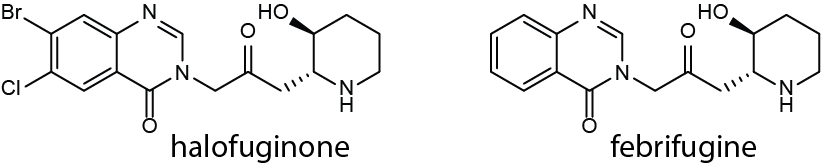
Halofuginone is one of the most potent antimalarials discovered to date. It is an analog of febrifugine, the active ingredient of a #TraditionalChineseMedicine herbal 🌿remedy, which has been used for millennia to treat fevers🤒and malaria. 3/
1
1
5