Linda B. Baughn
@lbaughn
Followers
593
Following
2K
Media
43
Statuses
427
Geneticist and research scientist at Mayo Clinic working to better understand, diagnose and treat B-cell malignancies. Opinions on Twitter are my own.
Minnesota
Joined April 2009
@theMMRF @MMRFTeam4Cures An incredible experience. 30 miles of hiking with new friends. The best part: the team raised $75K for MM research! So grateful for the opportunity @GD_251
1
2
13
Standardizing FISH testing in myeloma is critical to reduce lab to lab variability and misinterpretation. We propose guidelines for FISH panel design and reporting to align with the new IMS/IMWG risk stratification. @CG_Consortium @MayoMyeloma.
Just out: Guidelines for testing and reporting cytogenetic results in myeloma @BloodCancerJnl #OpenAccess Bookmark!. Allows you to incorporate latest IMS/IMWG high risk criteria in practice. @lbaughn @RahulBanerjeeMD @Rfonsi1
0
11
20
RT @VincentRK: We will also be providing recommendations to other labs on how to perform and interpret to meet the IMS/IMWG definition with….
0
3
0
RT @AnetaMikulasova: We’re hiring!.Postdoc in Computational Genomics at @EdinUni_IGC .Focus: myeloma genome, structural variants, heterochr….
elxw.fa.em3.oraclecloud.com
We are seeking a Postdoctoral Research Associate in Computational Cancer Genomics to join our team at the University of Edinburgh, funded by a Medical Research Council.
0
6
0
RT @JoachimSchork: Visualize genomic data with ease using gggenomes, an R package that extends ggplot2 to handle and display genomic inform….
0
124
0
We are eager to hear from the community clinicians!.
We have to reach physicians in the community to see how we can get their input. Can @ACCCBuzz help to get this survey out and partnering with them. Apart from risk stratification, can we report that BCL2 inhibitors can be used in the FISH report. @PedalheadPHX @lbaughn.
0
0
2
Thank you to those who have completed the myeloma FISH survey! We are so grateful. If you haven’t yet completed the survey, we ask you to please consider making your voice heard. Your responses are so valuable. #MMSM @RahulBanerjeeMD.
Our goal is to improve FISH reporting for myeloma. If you are a myeloma clinician, please consider this survey. Your responses will be helpful as we propose solutions for standardization and improve reporting clarity. Thank you! @MayoMyeloma.
0
2
9
RT @MarkARubin1: Grand challenge of moving cancer.whole-genome sequencing into the clinic. Comment on the largest whole-genome sequencing s….
nature.com
Nature Medicine - The largest whole-genome sequencing study thus far has revealed myriad actionable alterations and potential biomarkers for 33 cancer types, but various logistical, technical and...
0
34
0
RT @FrancescoMaura4: Excited to see our new paper entitled "Genomic Classification and Individualized Prognosis in Multiple Myeloma" out on….
0
54
0
Our goal is to improve FISH reporting for myeloma. If you are a myeloma clinician, please consider this survey. Your responses will be helpful as we propose solutions for standardization and improve reporting clarity. Thank you! @MayoMyeloma.
Are you a myeloma clinician who orders FISH tests? If so, the CGC Plasma Cell Neoplasm Working Group is interested in your feedback! FISH reports can be confusing and we want to learn about your experiences to help optimize cytogenetic reports. Survey:
1
14
28
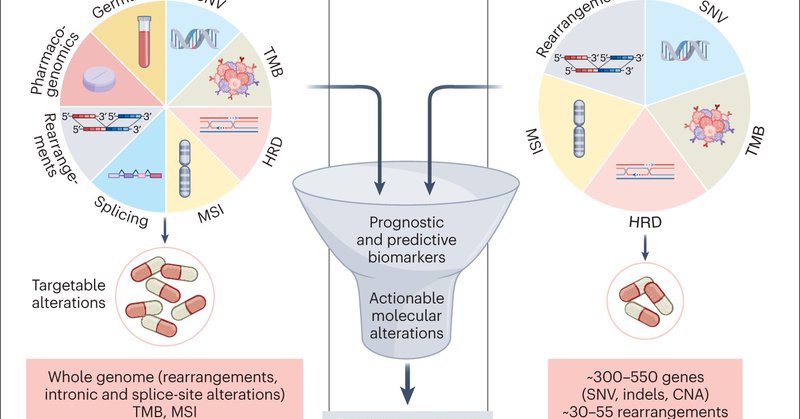
Unbalanced MYC rearrangements are found in about 12% of HGBCLs. We asked: do these represent real MYC rearrangements. Interpretation for a cytogeneticist is challenging.
nature.com
Blood Cancer Journal - Unraveling the genomic underpinnings of unbalanced MYC break-apart FISH results using whole genome sequencing analysis
1
2
4
RT @tangming2005: single-cell folks, what's the cell annotation tool do you recommend for immune cells and tumor cells? singleR? Seurat bas….
0
18
0
Thank you #IMS23 for giving us a chance to present our SNP rs9344 and t(11;14) FISH testing studies. Great to catch up with colleagues in person, initiate new collaborative opportunities and learn about advances in myeloma treatment. @MayoMyeloma
0
1
10
Lucky to have my poster next to @BioBenBarwick poster which definitely brought in a lot of foot traffic #IMS23
0
0
5