Kishwar
@kishwarshafin
Followers
1K
Following
987
Media
94
Statuses
557
@kishwar.bsky.social Research Scientist @Google. Interested in ML in genomics. @ucsc alumnus. 🇧🇩 🇺🇸
Mountain View, CA
Joined May 2016
It’s been an honor talking about our contributions to the pangenome project. Attached youtube video has the full story. Also this was a full circle moment for me.
Google Researchers worked closely with sciences in the Human Pangenome project to improve the pangenome using Google Research's DeepVariant and DeepConsensus methods, which use deep learning to improve the quality of genomics data. Learn more → https://t.co/qMi3xXiwnV
0
4
50
5/ Finally - some exciting progress applying AI in cancer research. Our C2S-Scale 27B foundation model, built with @Yale and Gemma, generated a novel hypothesis about cancer cellular behavior that was validated in living cells and we’ve released the model on GitHub and
21
34
349
Accurate somatic small variant discovery for multiple sequencing technologies with DeepSomatic https://t.co/i0nwOl1xBy
2
22
66
DeepSomatic, an AI model developed with @ucscgenomics, identifies cancer cell genetic variants. In research with Children’s Mercy, it found 10 variants in pediatric leukemia cells missed by other tools. DeepSomatic & the CASTLE dataset are now available: https://t.co/m0NVEdkam1
19
120
555
What if we could use AI to identify genetic variants in cancer cells? DeepSomatic uses ML to find key DNA variants with higher accuracy, aiming to create a better understanding of cancer’s underpinnings and support partners in designing more effective, personalized treatments.
12
46
279
For 10 years, Google has worked to accurately read the operating manual of all life on Earth — the genome. Our AI tools are now used by partners for real-world challenges from improving healthcare to biodiversity conservation. Check out the key milestones ↓
39
289
2K
DeepSomatic is out today in @NatureBiotech, demonstrating outstanding accuracy in all major sequencing platforms for somatic variant detection. Incredible collaboration between @GoogleResearch, @ucscgenomics, @ChildrensMercy, @NCICancerCtrl etc. https://t.co/ab7N4uuhRI
Today, @GoogleResearch announced DeepSomatic, a new machine learning model developed with our partners, including @ucscgenomics and @ChildrensMercy, that accurately identifies genetic variants in cancer cells — a critical step for delivering more precise treatments for patients.
0
4
18
Release of DeepVariant and DeepSomatic v1.9 DV: Added training on HG002 T2T-Q100. Error reduction of 12% for Illumina and 30% for PacBio on this truth set. 25% faster. DeepTrio is 5x faster (20h -> 4h). DS: New models FFPE_TUMOR_ONLY for {WGS, WES}. Much improved WGS models.
1
26
93
DeepVariant 1.8 is out. Pangenome is here and it just got ~50% faster.
Release of DeepVariant 1.8. Large speed improvement (~67% faster) via small model for easy sites. New Pangenome-aware option. Reduces error by ~30% for vg-mapped WGS ~10% for BWA WGS ~5% BWA exome. New config for custom model users, see release notes 1/3 https://t.co/TQmQElqAOR
0
12
77
Initiated in 2023 by @marianattestad and @_beenkim, this project has been a collaborative effort. For over a year, Atilla and Yuchen dedicated 20% of their time to working with @_beenkim , @acarroll_ATG and myself on this project. It has been a fun exploration!
When we train deep learning models for genomics, what do they learn? To help answer this question, we examined the DeepVariant model to determine what insights it has developed, and we discovered some surprising concepts embedded within. Read more at https://t.co/SSUh4EdJVo
2
4
13
Transformer-based polishing approach DeepPolisher's manuscript is now live. @miramastoras polished 180 assemblies using DeepPolisher for the next human pangenome release. Collaboration with @BenedictPaten @MobinAsri. PM @acarroll_ATG and eng mng @pichuan. https://t.co/lOmGI29ai1
biorxiv.org
Accurate genome assemblies are essential for biological research, but even the highest quality assemblies retain errors caused by the technologies used to construct them. Base-level errors are...
0
16
38
Release of DeepSomatic v1.7 ( https://t.co/YpB46RlP89). Now supports tumor-only applications, now supports FFPE-prepared samples with specific models. New models to support exome and ONT. Improved accuracy and runtime improvements.
github.com
DeepSomatic is an analysis pipeline that uses a deep neural network to call somatic variants from tumor-normal and tumor-only sequencing data. - google/deepsomatic
1
28
96
Finally, all of the models and improvements will be available in the next official DeepSomatic release with the documentation and a link to the manuscript. Currently, you can use the docker mentioned in the manuscript to test the models. 🧵7/7
0
0
0
Finally, we also extended DeepSomatic's ability to call variants with tumor-only data from WGS, PacBio and ONT sequencing. We also extended to FFPE_WGS and FFPE_WES for tumor-normal variant calling. 🧵6/7
1
1
2
We re-trained DeepSomatic with all cell lines and with data varying tumor-normal purities. The new models show significant improvements against the orthogonal truth sets generated to verify the improvement. 🧵5/7
1
0
0
However, the lack of training set in somatic space is a true bottleneck. @jiimiinpaark lead the work to develop a training set with five tumor-normal cell lines using three sequencing technologies. Massive effort to have more data in this space. All data is public. 🧵4/7
1
0
0
We trained our initial model on SEQC2 data ( https://t.co/DiIGaC4MaW) and showed DeepSomatic performs better than existing somatic variant callers. 🧵3/7
1
0
0
We developed DeepSomatic by making significant changes to the DeepVariant framework. Instead of calling germline variants with genotypes, we trained the models to classify somatic, germline or reference by representing both tumor-normal reads in the example. 🧵2/7
1
0
0
DeepSomatic ( https://t.co/BqZkDNMv1E) preprint is out showing improvements in somatic variant calling in various platforms. Work lead by @jiimiinpaark and @daniel_e_cook. Tumor-only lead by @pichuan. In collaboration with @acarroll_ATG, @MishaKolmogorov and @BenedictPaten. 🧵1/7
github.com
DeepSomatic is an analysis pipeline that uses a deep neural network to call somatic variants from tumor-normal and tumor-only sequencing data. - google/deepsomatic
DeepSomatic: Accurate somatic small variant discovery for multiple sequencing technologies https://t.co/l42tuwqoCh
#biorxiv_bioinfo
1
18
58
Our fast haplotagging paper is now published ( https://t.co/QcCE19MrQn) - see the earlier thread for a summary. Thanks to @kishwarshafin @AlexeyKolesni18 for leading the work and collaborators: @Johngorzynski, @gsneha261, @euanashley, @mitenjain, @khmiga, @BenedictPaten
nature.com
Nature Communications - DNA variant calling methods based on deep neural networks can use local haplotyping information with long-reads to improve genotyping accuracy, however this increases...
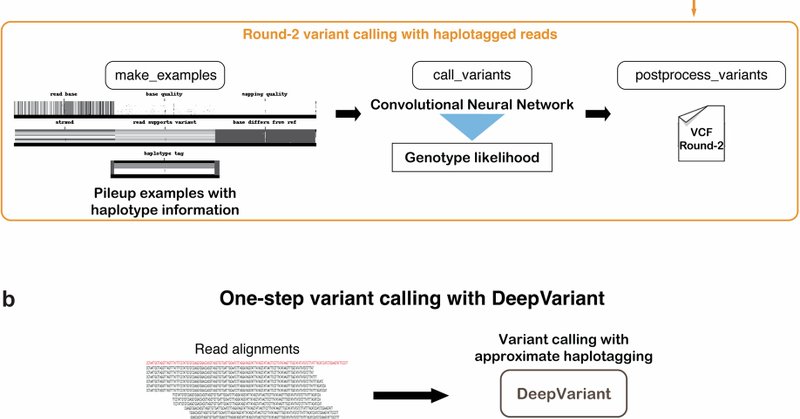
Happy to share this paper which provides more detail about the process we use to assign haplotypes to long reads on-the-fly, which enabled us to speed up the DeepVariant release at v1.4. Implementation by Alexey Kolesnikov, Lead for collaboration, writing, figures @kishwarshafin
0
13
43