Karen Miga
@khmiga
Followers
4K
Following
3K
Media
44
Statuses
2K
Associate Professor at UCSC; UC Santa Cruz Genomics Institute; Satellite DNA Biologist
Joined February 2014
RT @HumanPangenome: HPRC is building a reference that better reflects global genomic variation. We're well on our way to advancing assembl….
0
11
0
RT @HumanPangenome: 📢 HPRC Release 2 is here! . Now with phased genomes from 200+ individuals, a 5x increase from Release 1. Explore sequ….
0
20
0
RT @ArbelyYael: Very excited to share our work @MahlkeMegan in great collaboration with @NAltemose @khmiga Peter Campbell and Rachel O'Neil….
0
6
0
RT @humangenomeorg: Meet our plenary speakers!.We are pleased to announce the distinguished plenary speakers for HGM2025. Come and listen t….
0
4
0
RT @humangenomeorg: Final call to submit your abstract for the presentation during HGM2025! .Travel grants for students/post-docs and early….
0
3
0
RT @humangenomeorg: Meet our speakers!.We are pleased to announce the distinguished speakers for HGM2025. See you all at HGM2025!.Register….
0
7
0
RT @acarroll_ATG: Release of DeepVariant 1.8. Large speed improvement (~67% faster) via small model for easy sites. New Pangenome-aware opt….
0
57
0
RT @HumanPangenome: Accurate genome assemblies are key for research. DeepPolisher uses Pacbio HiFi alignments to improve base-level accurac….
0
16
0
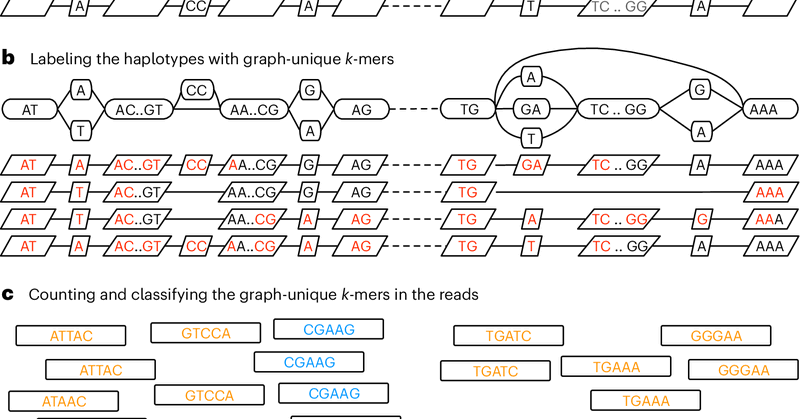
RT @HumanPangenome: Personalized pangenome preferences serve as crucial breakthroughs, furthering HPRC's mission to diversify genetic repre….
nature.com
Nature Methods - This work introduces a k-mer-based approach to customizing a pangenome reference, making it more relevant to a new sample of interest. This method enhances the accuracy of...
0
10
0
RT @mrvollger: I am going on the faculty job market this year, so if your home department is starting a search, I'd love to hear about it.….
0
9
0
RT @potapova_tamara: New preprint from our lab 🚨 We explored the variability in ribosomal DNA copy number and activity across humans of the….
0
25
0
RT @GoogleHealth: Genome sequencing is faster and cheaper than ever – but obstacles to broad adoption remain. Join @Google’s genomics lead….
0
18
0
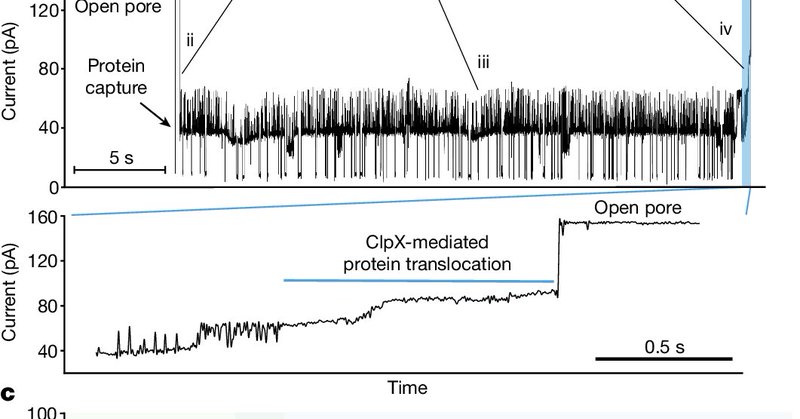
RT @jeffnivala: Published today in @Nature, we describe an approach for single-molecule protein reading on @nanopore arrays. By utilizing C….
nature.com
Nature - A technique for threading long protein strands through a nanopore by electrophoresis and back using a protein unfoldase motor, ClpX, enables single protein molecules to be analyzed...
0
290
0
A friendly reminder that today (August 1st) is the last day to register for the T2T-F2F! Hope you can join us (details below and reach out if you have any questions).
We welcome you to join us for the T2T Consortium Face-to-Face (T2T-F2F) Meeting in Santa Cruz, CA Sept 3-4. Open to all -- register before August 1st:
1
8
18
RT @acarroll_ATG: Our fast haplotagging paper is now published ( - see the earlier thread for a summary. Thanks to….
nature.com
Nature Communications - DNA variant calling methods based on deep neural networks can use local haplotyping information with long-reads to improve genotyping accuracy, however this increases...
0
13
0
RT @GenomeTDCC: 🧬Want to learn more about the Telomere-to-Telomere project and the folks involved in the T2T Consortium before the T2T-F2F….
0
2
0