Rasmus Kirkegaard
@kirk3gaard
Followers
3K
Following
26K
Media
1K
Statuses
11K
Staff scientist having fun with DNA seq and bioinformatics at #AlbertsenLAB. 🦋@kirk3gaard.bsky.social. COI : I am a @nanopore shareholder.
Joined August 2011
Want @nanopore data for benchmarking microbe related stuff? Check our collection of data for pure cultures, @ZymoResearch mocks and complex metagenomes ( https://t.co/EDtk6kQtKk). Basecalled reads with methylation is in @ENASequence. Eventually the pod5s might go there as well.
github.com
Overview of our data for microbial genomic benchmarking - Kirk3gaard/MicroBench
2
11
50
Roche SBX very high throughout NGS instrument is about to disrupt the short reads sequencing space. But is the data quality good enough? Read all about it at
1
3
10
Sorry but has nothing to do with @nanopore vs @PacBio but purely comes down to poor library preps... TLDR short reads are worse than long reads for assembly (regardless of the tech) 😉
New #metagenome assembly comparison of PacBio, ONT, and Illumina in gut #microbiomes. - "HiFi generated more cMAGs than ONT at every depth with either assembly method" - "HiFi cMAGs had significantly higher completeness and lower contamination than ONT" https://t.co/1BCNZd5qfC
1
1
10
Pretty cool. Illumina Protein Prep | Detect and quantify proteins with NGS
illumina.com
This automated NGS-based proteomics solution with SOMAmer technology and Illumina sequencing and analysis detects and quantifies 9.5K human proteins in 2.5 days.
0
2
7
"myloasm demonstrates that ONT and HiFi reads are becoming more comparable, with myloasm even assembling more circular and complete genomes with ONT than HiFi"
Preprint out for myloasm, our new nanopore / HiFi metagenome assembler! Nanopore's getting accurate, but 1. Can this lead to better metagenome assemblies? 2. How, algorithmically, to leverage them? with co-author Max Marin and supervised by Heng Li @lh3lh3
0
2
7
How many TBp/day of @nanopore basecalling can it do?
Europe’s first exascale supercomputer - JUPITER - is now live. Powered by NVIDIA Grace Hopper, @fzj_jsc's #JUPITER is the world’s most energy-efficient supercomputer, fusing AI and HPC to drive breakthroughs in climate science, neuroscience, quantum simulation, and more. 👉
0
0
6
🚀 New release of our ONT methylation Nextflow pipeline! Developed by my PhD student Valentina at RKI’s Genome Competence Center 🧬 🔧 Updates: - Updated Modkit envs - Auto-thresholding for confidence - Improved README Check it out 👇 🔗
github.com
Contribute to rki-mf1/ont-methylation development by creating an account on GitHub.
2
2
20
Illumina publishes a new "HP homopolymer Advanced Recipe" for the NextSeq 2000 instrument line.
1
3
10
@jjminich @kirk3gaard @nanopore I just tested H200, and got 2.88^7 samples per second. That's just under 200gb/day which is only slightly faster than the H100 tested at 197gb/day.. So yea, pretty similar.
2
1
2
@kirk3gaard @nanopore @PeterMacCC This was a larger test set, because it does your one too fast to benchmark the cooling 😅 4.9^7 samples/s So what's that...339.75GB/day? all for less than an H100 machine 🤣
1
1
6
I built and installed this absolute beast of a machine for @nanopore basecalling @PeterMacCC Threadripper 4x5090, liquid cooled 128gb DDR5 ram ~40Tb of nvme and SSD storage TWO power supplies, a 1200W and a 2500W All in a Corsair 9000D case ~5^7 samples/second with dorado.
7
5
73
To celebrate that ISME20 is exactly one year from now, the ISME20 Local Organising Committee recreates the magic of their first video by taking you on a new adventure in the second installment. Watch the brand new video: https://t.co/yQIppeAF8f
#isme20
0
4
14
News corp Just dropped part of their FSD Australia video. https://t.co/4WhZESJClQ
13
12
119
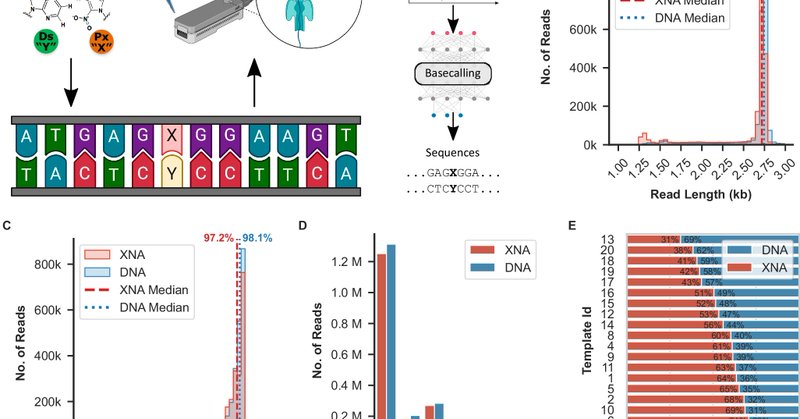
Our work on direct @nanopore sequencing of non-canonical bases in now out in @NatureComms! Read all about it here: https://t.co/fFl4Yncf83 Great collab with @ChewWeiLeong and Hirao labs
nature.com
Nature Communications - Perez, Kimoto, Rajakumar and colleagues present a fast and accurate DNA sequencing method that reads canonical and non-canonical bases using AI and nanopore technology. The...
Tech Alert! 🚀🧬 We can now determine the sequence of DNA with non-canonical bases in a direct and high-throughput manner with Nanopore sequencing. Check out our preprint for details:
0
9
24
High throughput method for monitoring SARS-CoV-2 variants in wastewater by Nanopore sequencing https://t.co/ZfqTl17I2F
0
5
4
skani v0.3.0 is released. https://t.co/dEkIzxIbDr * 30-40% potential reduction in memory * Breaking changes to indexing and searching databases Calculate ANI for contigs, genomes. Search vs > 140k genomes: pre-indexed GTDB-R226 available for download.
github.com
Fast, robust ANI and aligned fraction for (metagenomic) genomes and contigs. - bluenote-1577/skani
0
23
54
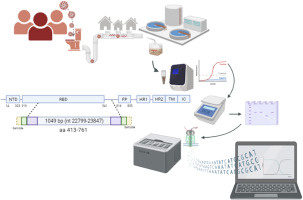
Interlaboratory evaluation of high molecular weight DNA extraction methods for long-read sequencing and structural variant analysis | BMC Genomics | Full Text
bmcgenomics.biomedcentral.com
Background Long-read sequencing technologies enable resolution of structural variants (SV) and long-range genome assembly, but require high molecular weight (HMW) DNA of both high quantity and...
0
2
9
Hopper is back 🚀 Dorado update from @nanopore adds some speed. Anyone happen to have access to the Blackwell server grade GPUs or are they all busy making silly ai images? https://t.co/5hcUn0uSeK
2
3
31
Open hardware desktop 3D printing is dead - you just don't know it yet ➡️
josefprusa.com
How Chinas strategic focus on 3D printing and patent incentives since 2020 created a "minefield".
6
41
201
@CatharineAquino Our protocol has been: guess the concentration from qubit/E-gel. Load iseq (with extra phix if needed). Use results to back calculate actual concentration (and rebalance if needed), then send out for higher depth sequencing.
0
1
5