Jacob Wardman
@jacobwardman1
Followers
212
Following
3K
Media
23
Statuses
73
Postdoctoral Fellow at @AstraZeneca. Exploring new frontiers in protein science a few mutations at a time. PhD from @withlabb at @UBC.
Joined January 2019
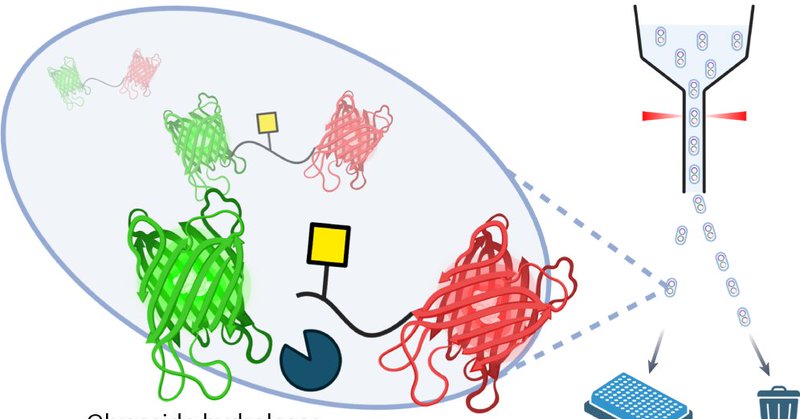
Very excited to have this paper come out in @nchembio! We develop and demonstrate a new methodology for characterizing and engineering enzymes active on mucin-type O-glycoproteins! .
nature.com
Nature Chemical Biology - By developing a genetically encoded biosensor for enzymes that act on O-glycoproteins, Wardman et al. have provided a new method for rapid screening and analysis of these...
5
12
86
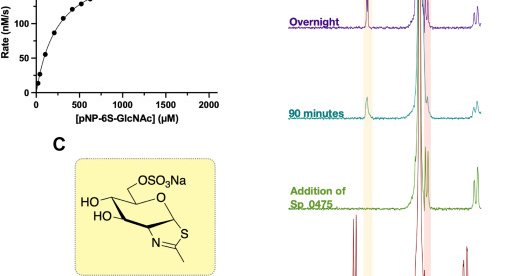
RT @rheabains: Excited to share that our work on sulfated oligosaccharide and glycan synthesis is now live in @J_A_C_S ! Check it out https….
pubs.acs.org
Sulfation is a common, but poorly understood, post-glycosylational modification (PGM) used to modulate biological function. To deepen our understanding of the roles of various sulfated glycoforms and...
0
11
0
RT @tomaashby: The Simpsons on PhD students - almost twenty years old & more cutting than ever
0
3K
0
RT @EricDLombardi: My latest for @TheHubCanada . CANADA CAN NO LONGER AFFORD ITS ‘SOFT CORRUPTION’ PROBLEM. While I’m wearing a tinfoil hat….
thehub.ca
0
174
0
I'm undoubtedly a bit late to the party but this is an incredible article from the incomparable @MaxGantz_ ! A great way to examine epistatic interactions at ultrahigh-throughputs. Congrats to all involved on such exciting and innovative work! .
biorxiv.org
Engineering enzyme biocatalysts for higher efficiency is key to enabling sustainable, ‘green’ production processes for the chemical and pharmaceutical industry. This challenge can be tackled from two...
1
0
3
I am excited to be selected for this program @AstraZeneca! It's a great privilege to be able to pursue my own idea and especially one that could help patients in the future. The challenge itself was a tremendous experience. Looking forward to the next steps!.
Eight early-career scientists from around the world have won the R&D Postdoctoral Challenge. The winners have been awarded postdoctoral positions at one of our strategic R&D centres to turn their own innovative research ideas into reality:
3
0
18
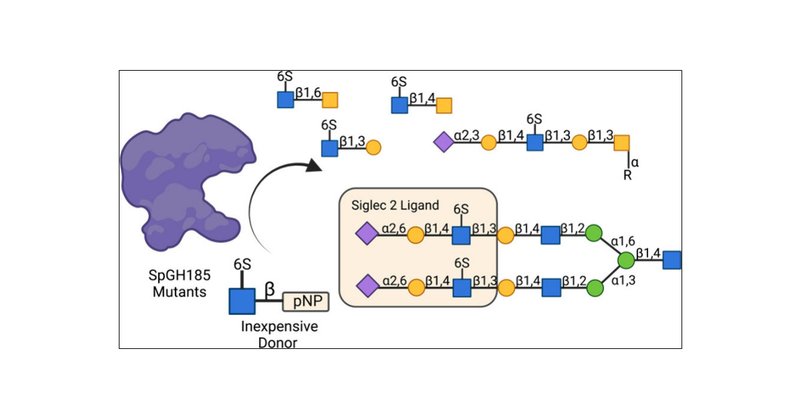
RT @rheabains: Exciting news! Our latest research on characterizing a new GH family is now published in @jbiolchem ! Huge thanks to my amaz….
jbc.org
Sulfation is widespread in nature and plays an important role in modulating biological function. Among the strategies developed by microbes to access sulfated oligosaccharides as a nutrient source is...
0
16
0
Of course, this wouldn't have been possible without the incomparable @wakarchuk, @boraston_lab, @lya_sim, and many others + the wonderful environment of @ubcmsl @UBCbiochemistry @UBCChem.
0
0
1
To do so, we made a fluorescent protein FRET pair. We then used engineered E. coli (part of a longstanding collab with @wakarchuk) to glycosylate the linker between the proteins by introducing specific seqs. This glycosylation occurs in vivo with low cost + high yields
1
0
0