Matthias Heinig
@heinig_matthias
Followers
190
Following
23
Media
17
Statuses
51
Joined February 2018
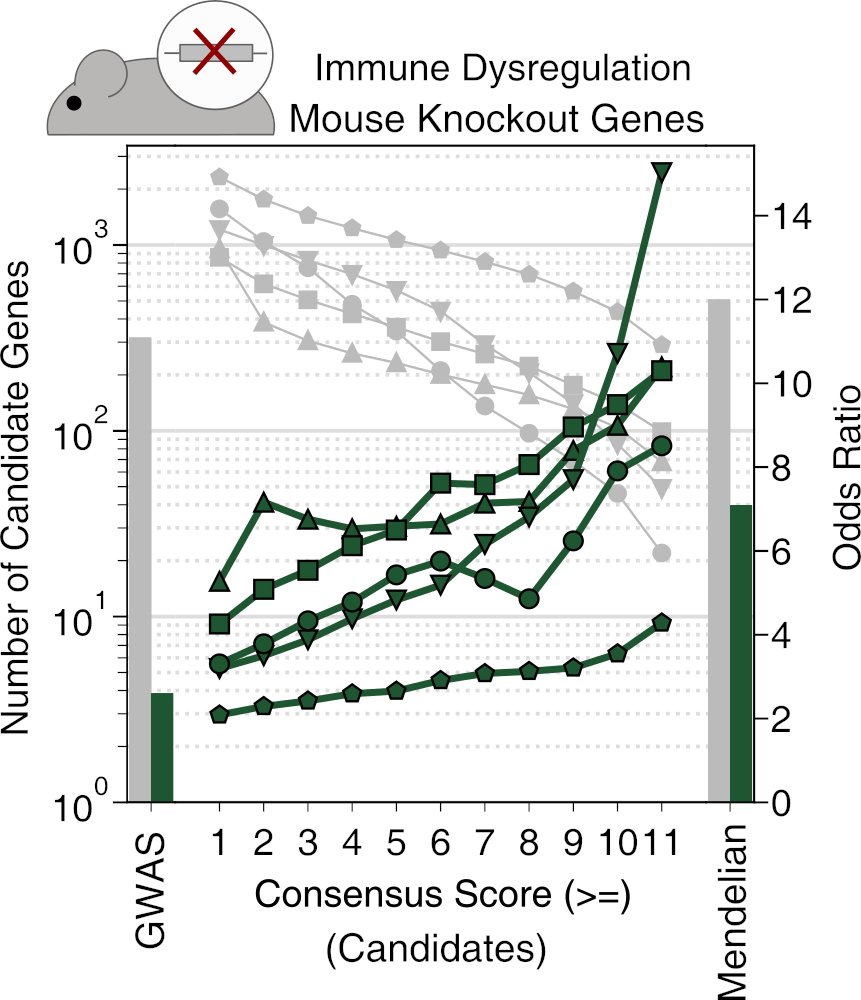
9/9 A huge thank you to our fantastic collaborators @Jessica_Pauli_ , @NadjaSachs, @LarsMaeg from @TU_Muenchen and @_MatthiasMunz, Ehsan Vafadarnejad, Tania Carrillo-Roa, Peter Kastner from @Roche! @CompHealthMuc.
0
0
3
1/9 Excited to share our new preprint! Led by @KorbinianT, we present the Integrated single-cell Atlas of Human Atherosclerotic Plaques—a resource with 261,747 annotated cells from carotid, coronary, and femoral arteries.
1
5
11
RT @KPekayvaz: 1/Thrilled to share our most recent efforts @NatureMedicine on joint multi-omic analyses unravelling the comprehensive immun….
0
10
0
RT @gagneurlab: Our study on genomic and transcriptomic aberrations over 3,700 leukemia samples is now published! AbSplice on cancer, driv….
0
7
0
This was a great collaboration between @KPekayvaz @nicolai_leo and Konstantin Stark @LMU_Uniklinikum and @Corinna_Losert @CompHealthMuc @HelmholtzMunich.
1
0
4
7/7 Super happy about this great work by @FlorinRatajczak and a great team effort with @interactome @HmguInet @HelmholtzMunich @CompHealthMuc.
1
0
5