Explore tweets tagged as #Intogen
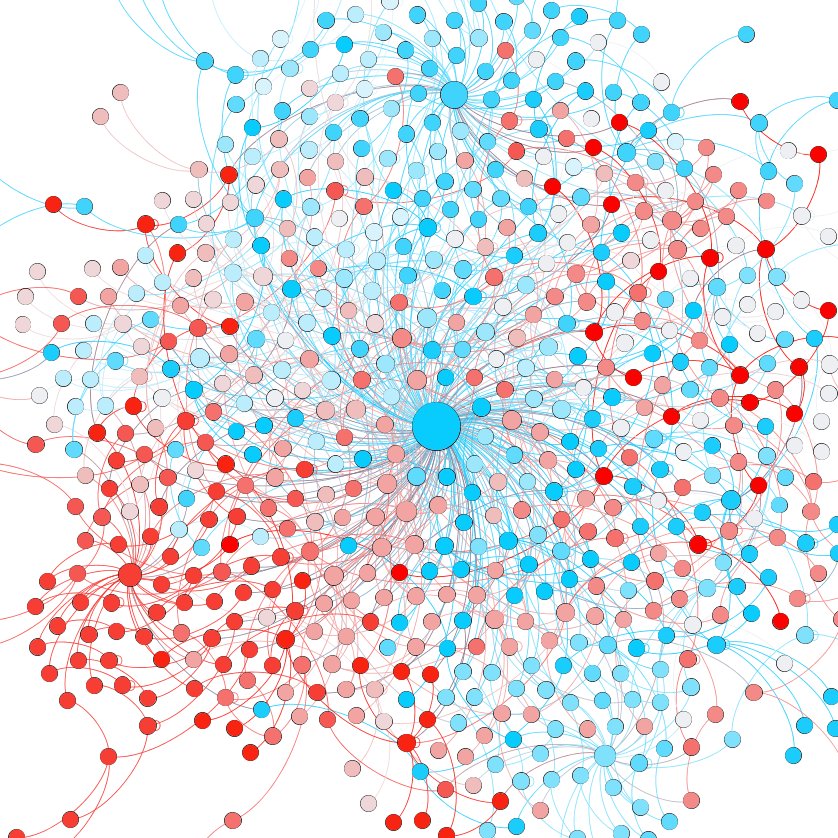
Using InPheRNo, we can identify cancer-specific TRNs. Below is a #BreastCancer specific TRN subnetwork, where transcript factors are the red nodes and the genes they regulate is green. Nodes with a blue border are listed as BRCA drivers by IntOGen and DriverDBv2.
1
0
1
First, cancer driver genes. We used the fantastic intOGen from @bbglab to detect cancer driver genes in each variant call set. As you can see, depending on which variant call set you use, you get very different results.
2
4
9
Conference time! First up @nlbigas telling us about intogen and boostDM which aim provide a compendium of mutations in cancer by screening all mutations in silico to determine “driverness” of mutations per cancer and per gene. #EACRbioinformatics #EACRvirtualevents
1
2
11
A compendium of mutational cancer driver genes.By @nlbigas & colleagues .They discuss the Integrative Oncogenomics (IntOGen) platform of annotating cancer driver mutations. Eg: “Mutation cloud” from ~900 #PancreaticCancer cases👇🏽.
0
13
30