Explore tweets tagged as #BioNumPy
See, the word 'vanilla' is actually used... Also, take a look at BioNumPy package, it might be nice.
1
0
0
BioNumPy: array programming for biology @naturemethods • BioNumPy revolutionizes biological data analysis by integrating the power of NumPy-like arrays, making Python even more accessible to bioinformaticians. • It enables direct handling of biological formats (like FASTQ,
0
37
164
BioNumPy: Fast and easy analysis of biological data with Python https://t.co/Nm0FVvxcdr
#biorxiv_bioinfo
1
63
273
BioNumPy: array programming for biology. Now it is on Nature Methods! BioNumPy package, which enables efficient and intuitive array programming on biological data in Python. https://t.co/f9DpFg956f
0
1
10
#BioNumPy: a #Python library built on top of #NumPy, for enabling array programming on biological datasets. It aims to make it easy to read common #bioinformatics file formats efficiently into NumPy-like data structures that enable efficient operations and analysis of the #data.
2
7
20
NumPyの上に構築されたバイオインフォマティクス用のライブラリBioNumPyが公開されました。githubのレポジトリーにはさまざまな使い方・チュートリアル記事も置かれています。 https://t.co/uc5QgoSolU
0
34
236
0
0
0
BioNumPy: Fast and easy analysis of biological data with Python. #SequenceData #Python #Bioinformatics @biorxiv_bioinfo
1
64
292
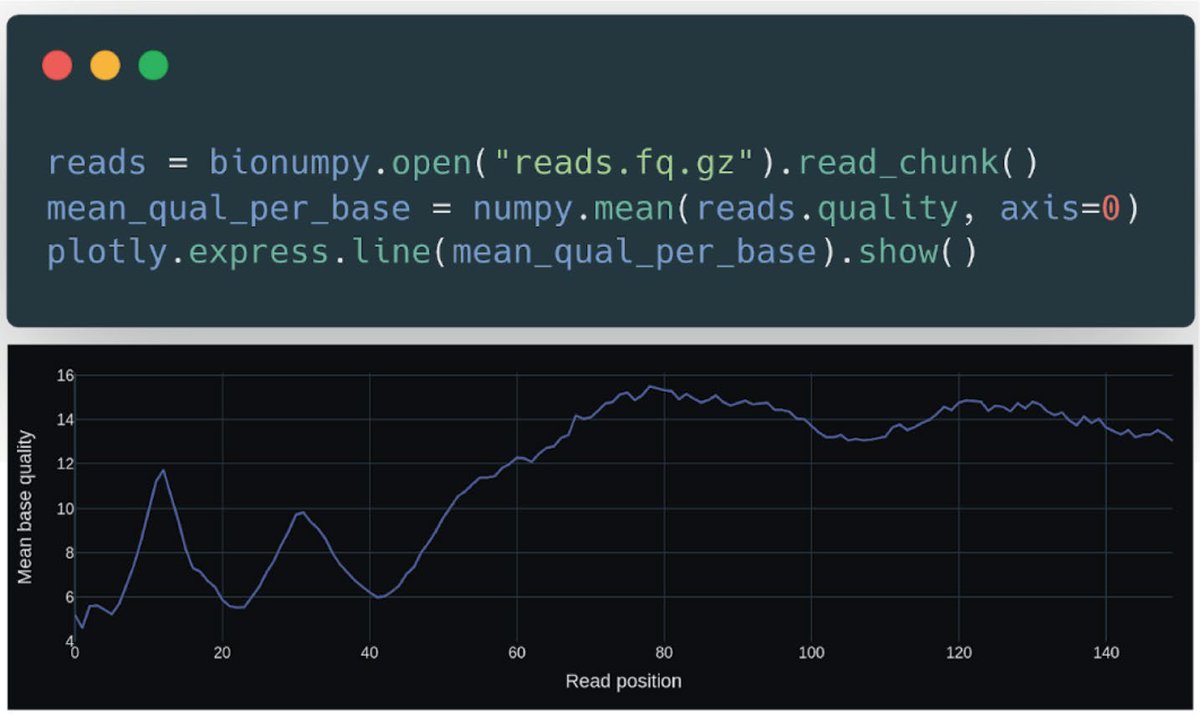
BioNumPy is able to efficiently load biological datasets (e.g. FASTQ-files, BED-files and BAM-files) into NumPy-like data structures, so that NumPy operations like indexing, vectorized functions and reductions can be applied to the data.
3
19
195
It’s quite easy to e.g. find the mean read pileup value around transcription start sites or other locations.
1
0
1
.. or around transcription factor peak summits. Here we plot the pileup for reads on the positive and negative strand, and clearly see the pileups we expect around the summit.
1
1
2
The Genome object makes it easy to read bam-files, bed-files and other data that belongs on a reference genome. Getting a pileup from intervals and slicing the pileup can be done with NumPy-like syntax.
1
0
1
BioNumPy: array programming for biology. Now it is on Nature Methods! BioNumPy package, which enables efficient and intuitive array programming on biological data in Python. https://t.co/qpBH3JnfQi
0
0
0
Discover how BioNumPy enhances biological data analysis by integrating NumPy's array programming with bioinformatics tools. https://t.co/dbvyi3irHl
0
0
5
Meet “BioNumPy”: A Powerful Tool for Quick and Easy Biological Data Analysis with Python https://t.co/nf0HzcPtIV via @CbirtDirector
#Bioinformatics #NumPy #Biology #data #BigData #DataAnalytics #Python3 #pythonprogramming #IoT #Genomics #scicomm #100DaysOfCode #AcademicTwitter
0
6
22
@vsbuffalo BioNumPy has some support for gtf files, but have not yet defined a complete set of abstractions for them. Would love suggestions for useful methods!
0
0
0