Francesco Andreace
@frnandreac
Followers
89
Following
278
Media
13
Statuses
33
PhD Candidate ALPACA 2020 ITN Sequence Bioinformatics @institutpasteur Paris Human Pangenomics Views are my own
Joined May 2022
It was nice to give a little contribution to this k-mer project :)
0
0
8
I don't want some AI bot that codes, does art, writes music or a book. What I want is a robot that cleans my damn house so I can have more time to do those things myself.
1
2
47
New preprint alert! 🚨 Our latest work on plasmid mobility with @epcrocha & @NucciAmandine is now available. Identification of novel origins of transfer across bacterial plasmids. Read the full paper here: https://t.co/Ww8KuXyNZf Thread🧵below!
biorxiv.org
Conjugative plasmids are important drivers of bacterial evolution, but most plasmids lack genes for conjugation. It is currently not known if the latter can transfer because origins of transfer by...
7
44
114
Honored to share an extraordinary day with Dr.Anthony Fauci! His insights on science, public health, and education inspired us.The engagement with doctoral students was a highlight, showcasing the incredible potential of the next generation in shaping the future of global health
0
7
26
Two days ago I had the honor to take a coffee with Dr Anthony Fauci before his keynote lecture on 40 years of HIV at @institutpasteur, together with other 9 selected phd students and postdocs. It was a privilege to ask him questions about policymaking and pandemic preparedness
0
1
4
Pangenome graphs are a powerful tool: choosing the right one for your application is paramount. Have a look at the characteristics of state-of-the-art tools! The reviewed article is now published on Genome Biology!
genomebiology.biomedcentral.com
Background As a single reference genome cannot possibly represent all the variation present across human individuals, pangenome graphs have been introduced to incorporate population diversity within...
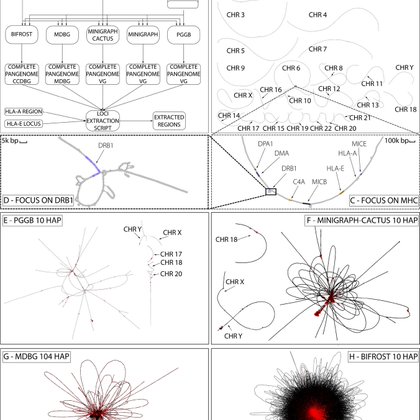
The construction of large cohorts human pangenome graphs is a challenging task. We selected 5 different state-of-the art tools that generate variation graphs or De Bruijn graphs and used them to construct a human pangenome of 104 high-quality human haplotype assemblies. (1/4)
0
6
18
Super thread by @camiladuitama1 on her PhD journey and the metagenomics methods she developed! If you are interested in ancient metagenomics contact her! She is super cool and can give you nice books recommendations as well!
5
0
3
We then summarized the strengths and the current bottlenecks of each graph-building tool based on different features to inform end users of the best approach for specific applications. #pangenomics #Health #genomics Thanks to @RayanChikhi, @yoann_dufresne and Pierre Lechat. (4/4)
1
0
4
We then examined differences in the topology of the graphs and how they represent variation, focusing of two HLA loci. HLA-E: a simple and relatively conserved locus and a complex region comprising several loci, including the HLA-A and the HCG4B genes. (3/4)
1
0
3
We constructed these graphs using PGGB, Minigraph, Minigraph-Cactus, Bifrost and mdbg without any preprocessing or chromosome separation to test the scalability and the stability of the tools under the same conditions. (2/4)
1
0
5
The construction of large cohorts human pangenome graphs is a challenging task. We selected 5 different state-of-the art tools that generate variation graphs or De Bruijn graphs and used them to construct a human pangenome of 104 high-quality human haplotype assemblies. (1/4)
Construction and representation of human pangenome graphs https://t.co/S7as2LjUSa
#bioRxiv
1
40
75
With plenty of colleagues, we are happy to introduce bioconvert, a software to convert bioinformatics files from one format to another. bioconvert currently includes 50 formats and 100 converters.
13
175
662
Spending millions of $, Grail created a TCGA-like study to systematically answer the following: What is the best cell-free DNA method to detect cancer from blood? 17 things we learned.🧵🩸
2
35
160