Francisco M Martín-Zamora
@fmmartinz
Followers
423
Following
1K
Media
71
Statuses
763
Scientist II, Computational Biology @altos_labs. Previously: PhD @Chema_MD, BSc @AlegreCebollada. London🇬🇧 | Madrid🇪🇸. Views my own. He/him🏳️🌈
London, England
Joined August 2019
A dream come true 😭❤️. My first-author paper is now out in @Nature .
21
66
587
RT @ArnauSebe: We offer three new positions to join BCA @MooreFound:.1. A full-stack developer to work on the BCA database with @emblebi.2.….
0
30
0
Truly excited to finally see this out! 👏🏻. Jorge's lab is where I started my scientific career, so this one's quite special. What a pleasure to have had a (very) small contribution to this article. Must read if you care about mechanosensing! 👀💡. Congrats to the whole team! 🎉.
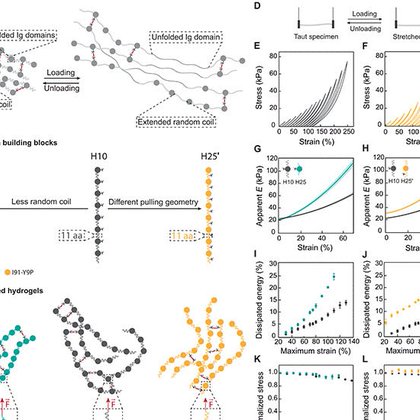
🎉🎉.Very happy that our paper “Cell response to extracellular matrix viscous energy dissipation outweighs high-rigidity sensing” is out @scienceadvances. A thread on the main findings follows👇. @CNIC_CARDIO @SBEsp @SEBBM_es.
0
0
4
RT @AlegreCebollada: 🎉🎉.Very happy that our paper “Cell response to extracellular matrix viscous energy dissipation outweighs high-rigidity….
science.org
Viscosity blunts cell response to stiff matrices in agreement with a pull-and-hold model of molecular clutch-based mechanosensing.
0
15
0
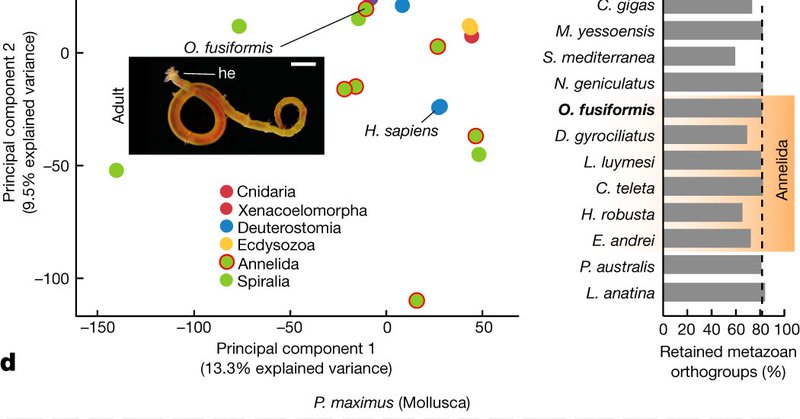
RT @a_pposada: We added Owenia fusiformis to our comparisons –a protostome thanks to which we can assess how much the deuterostome LCA chan….
nature.com
Nature - Comparative chromosome-scale genome sequencing and transcriptomic and epigenomic profiling of three different species of annelid provide insight into the evolutionary origin of larvae.
0
2
0
RT @a_pposada: I am very happy to announce that our work on hemichordate gene expression and chromatin accessibility dynamics is now publis….
0
27
0
RT @k_guynes: Another piece of work from my very talented friend - glad to have been part of this ☺️.
0
2
0
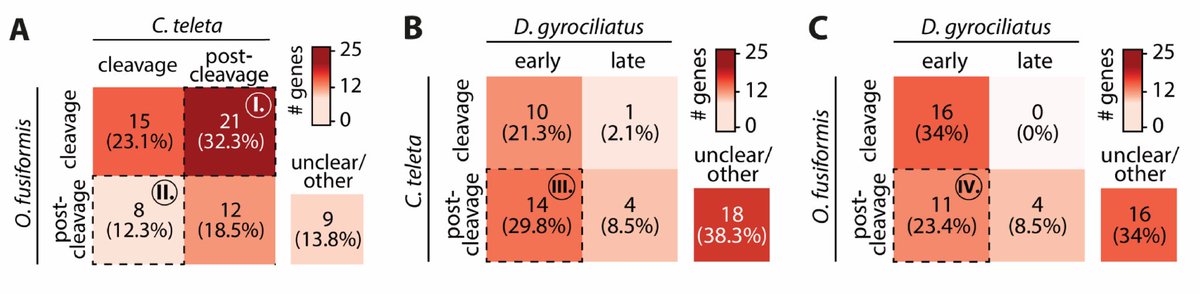
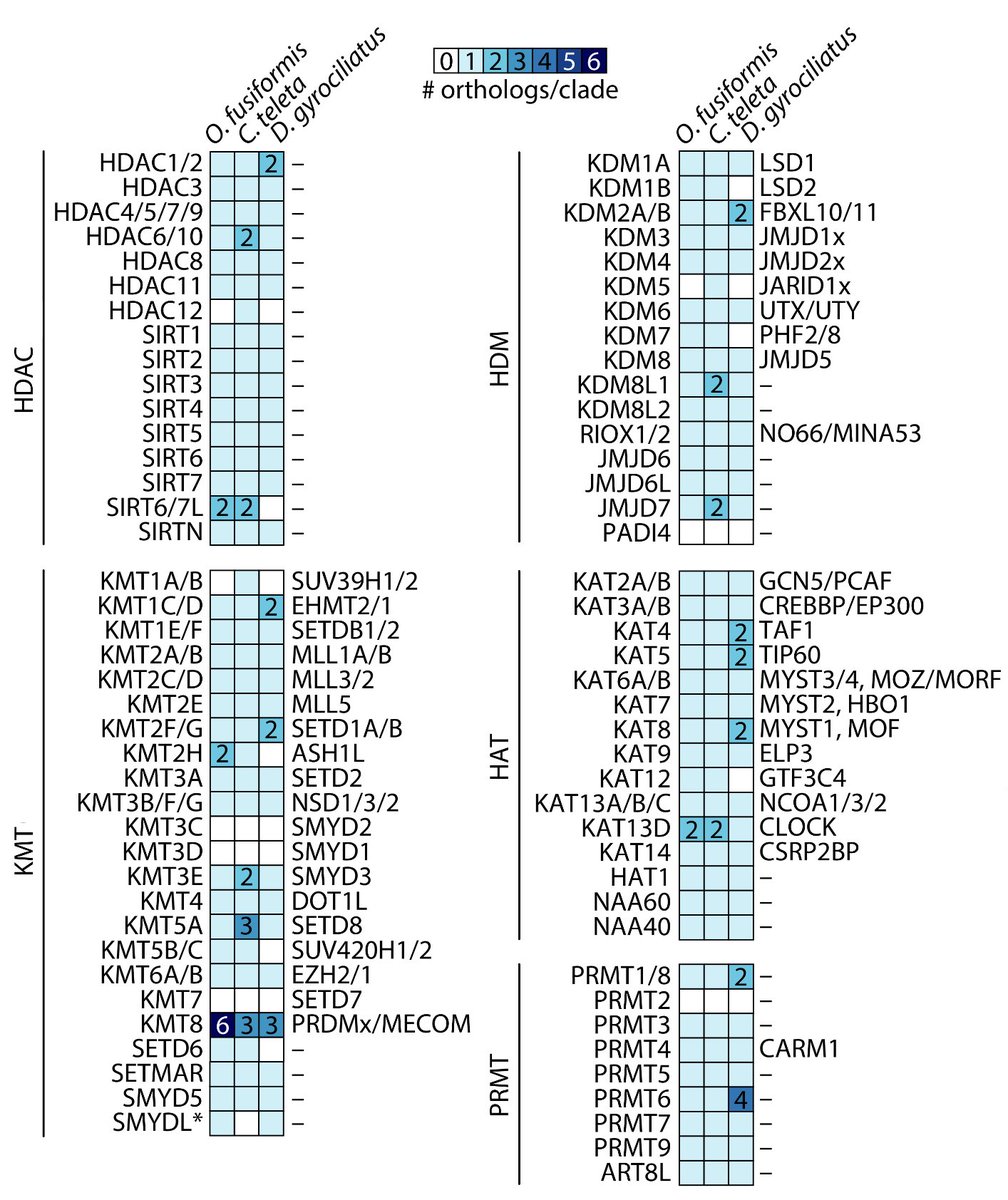
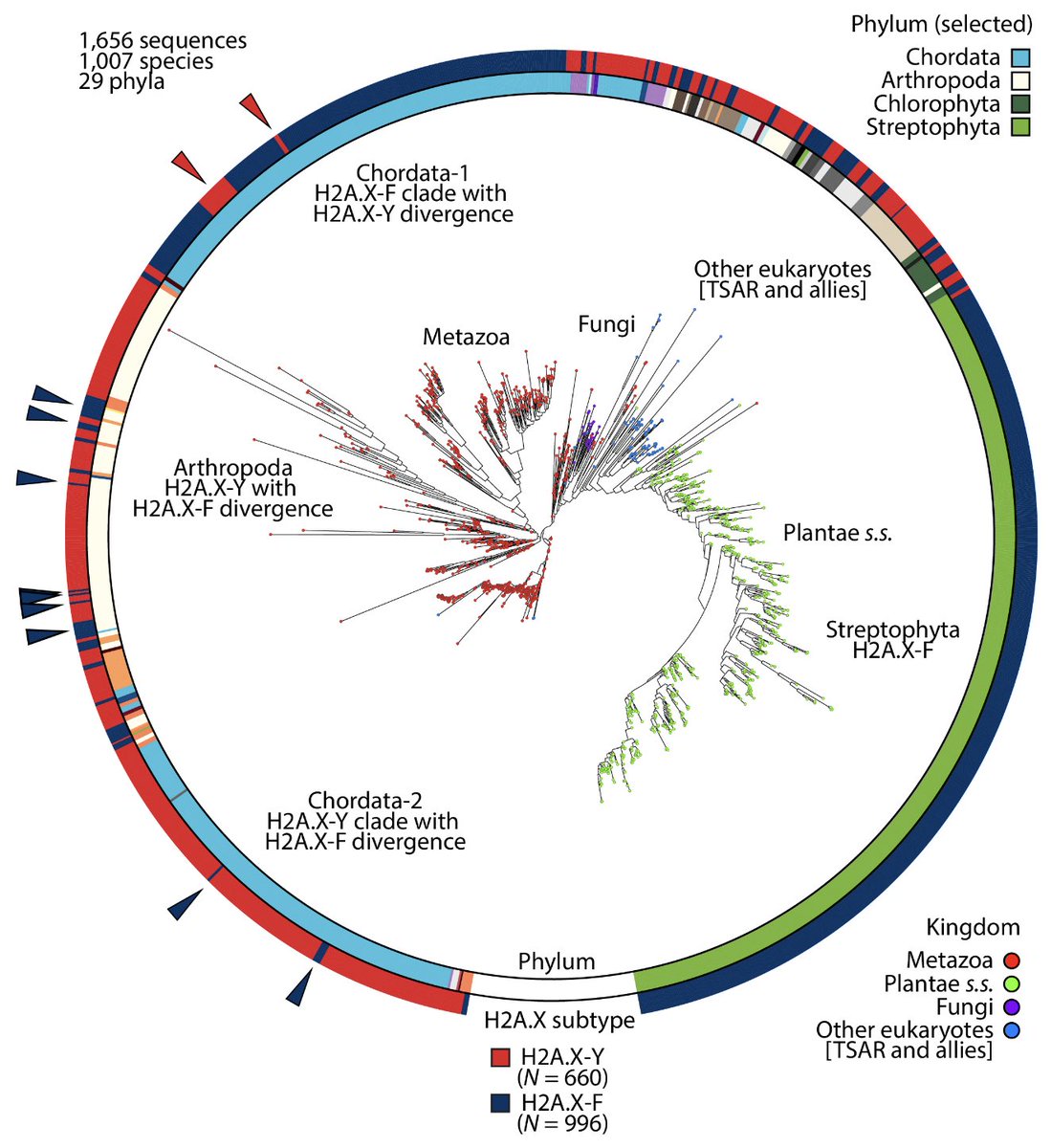
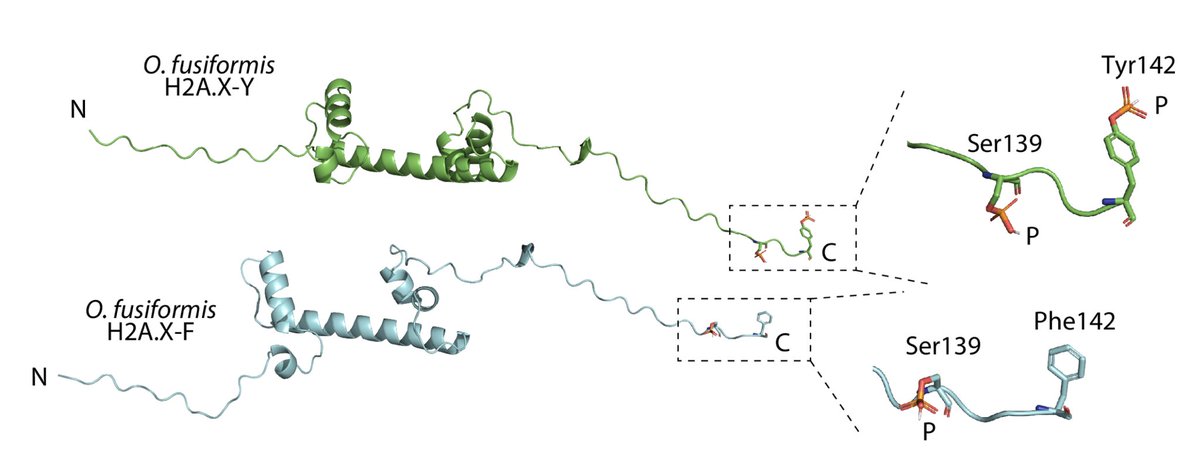
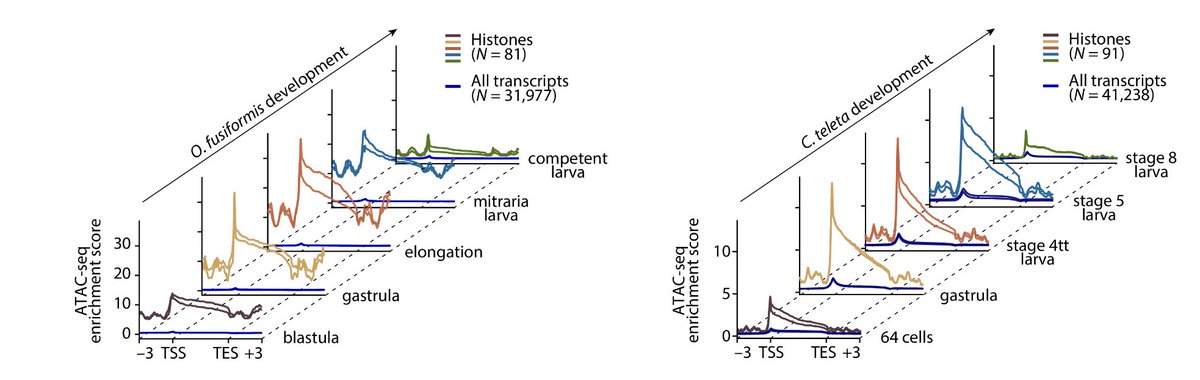
RT @Chema_MD: New paper from the lab! 🎉 Incredible effort by @fmmartinz to kick-start our work into annelid 🪱 histones and their regulatory….
0
10
0
I’m incredibly grateful to all those who helped get this story out. Firstly, to @Chema_MD and @pauljhurd for their guidance and mentorship, but also to @Rory_Donnellan7, @k_guynes, @Allan_littlecar, @Dr_Joby_Cole and Mark for their hard work 💪🙌. Thank you, truly! 🙏.
1
3
7