Josh Elias
@eliaslab1
Followers
492
Following
703
Media
8
Statuses
150
CZ Biohub Mass Spectrometry Platform Leader.
Stanford, CA
Joined December 2018
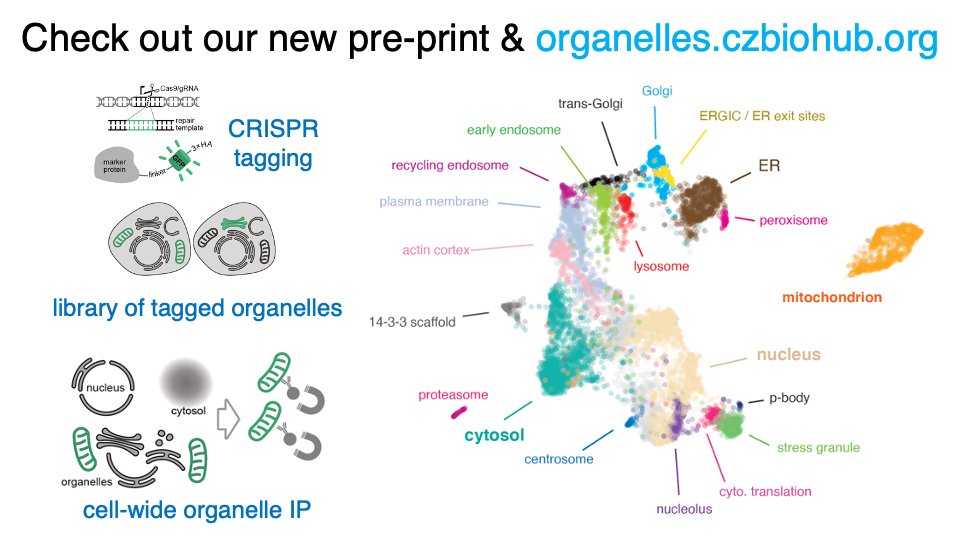
This is what interdisciplinary research looks like. What a pleasure to be part of this amazing team. Stay tuned! #organelleIP #czbiohub.
So stoked to be sharing this! New proteomics map of human subcellular architecture. CRISPR + organelle IP mass spec at scale. Check out pre-print and 🙌 to fantastic team @czbiohub SF & collabs! 1/2
0
8
20
RT @WeingartenShira: In a new study, we provide genome-wide view of #SARSCoV2 peptides that are presented on the HLA-II complex. We expose….
0
49
0
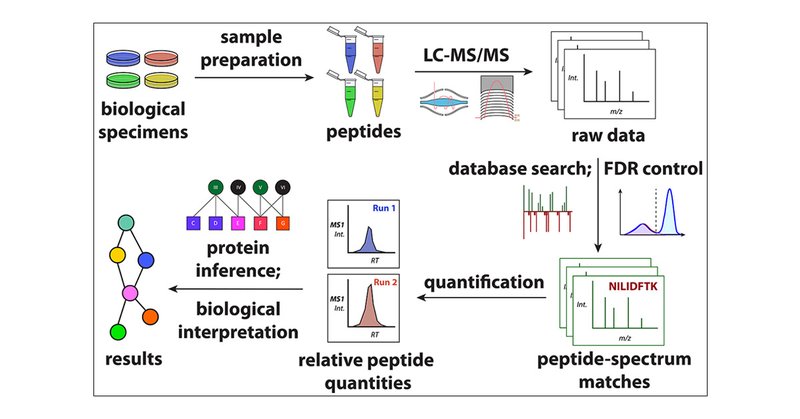
#TeamMassSpec-- how often do you get the FAQ: Can you please point me to a good primer on proteomics? @stshuken (now a @GygiLab postdoc) just published this thorough yet approachable solo author paper in JPR that should scratch this itch.
pubs.acs.org
Mass spectrometry is unmatched in its versatility for studying practically any aspect of the proteome. Because the foundations of mass spectrometry-based proteomics are complex and span multiple...
0
25
66
#TeamMassSpec — have you received this cockamamie garbage when trying to get replacement parts for your slightly older Thermo instruments? Pictured is our replacement ion transfer tube next to the original ‘V’ version. Purchased from the shop:
5
2
18
RT @kennethtodd: ChatGPT isn’t the only AI-powered website…. Here are 14 AI websites that feel illegal to know about:.
0
5K
0
RT @mateosfo: Do Menlo Park homeowners really need more power to keep “lowlifes” like … teachers and nurses and baristas from living anywhe….
0
1
0
Great to work with @MikeChuckLanz and @JanSkotheim on this fun project!.
We recently described how increasing cell size remodels the proteome and promotes senescence in cultured mammalian cells (Mol Cell's current issue). Here is some follow-up work on the same topic - mTOR and the cellular growth rate are central to the story
0
0
5
I was honored to participate in my last @Stanford PhD student’s graduation today. Dr. Marlene Heberling faced down far too many challenges to reach this day, and I am beyond proud of all she’s done and all she’ll do.
0
1
29
Hey #ASMS2022 job seekers! Torn between the basic research focus of academia versus team-driven science (and salaries) of industry? Don’t be. Come find @BrianCDeFelice or me to learn about job opportunities at @czbiohub We’re hiring at many levels.
czbiohub.org
Find job openings at the CZ Biohub Network.
0
6
21
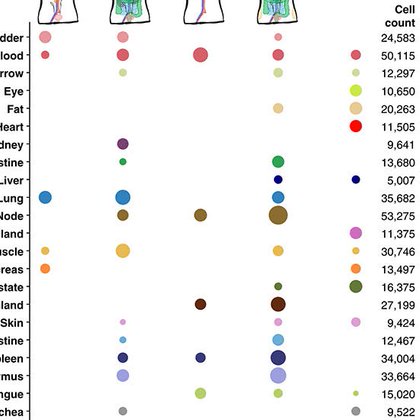
RT @czbiohub: Out today in @ScienceMagazine, the Tabula Sapiens is 1 of 4 new open-access cross-tissue human cell atlases. This is a major….
science.org
Single-cell transcriptomics provides a molecularly defined phenotypic reference of human cell types that spans 24 human tissues and organs.
0
63
0
RT @slschmid_CZB: Group Leader position open @CZBiohub for those interested in the systematic analysis of virus-host interactions and viral….
apply.workable.com
0
15
0
RT @michal_bassani: Join us for the HUPO-HIPP webinar and don't forget to apply to the coming HUPO-HIPP summer school in Oxford!.
0
8
0
Great work @LeonettiManuel and team! Can’t wait to see what happens next!.
Our paper is out today in @ScienceMagazine, I’m stoked @OpenCellCZB !. Check out CRISPR tagging + 3D live imaging + IP mass spec to map the human proteome. 👇 tweetorial from our 2021 pre-print.
0
0
3
Nice writeup of our collaboration with Chaitan Khosla @Stanford_ChEMH is moving the needle in #celiacDisease research. .
New research from Biohub and @Stanford offers a new window into #celiac disease and human digestion, shows promise for testing and drug development: .
1
2
15
Happy to report that our gluten urinary peptidomics paper is now out in Nature Communications: Retweeting @bpalanski 's excellent summary here. We hope it will usher more sensitive and accurate ways for managing celiac disease.
After a long week, thrilled to share the preprint of the last of my PhD projects from @StanfordUChem is posted👇 ! We developed a new #massspec method to analyze urinary peptides from patients with #celiac disease. A 🧵 (1/n)….
0
0
10
RT @EdHuttlin: If you’re new to Mass Spec - or even if you’re not - check out these videos from past years of the Mass Spec Summer School.….
0
11
0
Antigen presentation by MHC-2 is not simply a matter of binding affinity. In this collaborative work with Betsy Mellins, we show that large portions of MHC-2 repertoires are sensitive to the variable activity of the antigen loading Dm chaperone and it’s antagonist, Do.
Olsson et al.'s findings have implications for designing improved HLA-II prediction algorithms and research strategies for dissecting the variety of functions that different APCs serve in the body — @czbiohub @eliaslab1 @StanfordMed #TeamMassSpec #HLA.
1
5
24
RT @drAOPisco: We are looking for a Data Scientist to work across teams integrating mass spectrometry data with oth….
apply.workable.com
0
3
0
RT @slschmid_CZB: The CZ Biohub Network is scaling the successful @CZBiohub model to pursue the toughest, most important scientific challen….
0
19
0