dadejuan
@david_de_juan
Followers
109
Following
49
Media
0
Statuses
62
Joined January 2019
EL primero congreso de #congresosebibc2024 ya está aquí! Será del 16 al 18 de octubre de 2024 en Feria Valencia. La recepción de abstracts ya está abierta, apunta la fecha e inscríbete! Seguiremos actualizando próximamente.
1
40
61
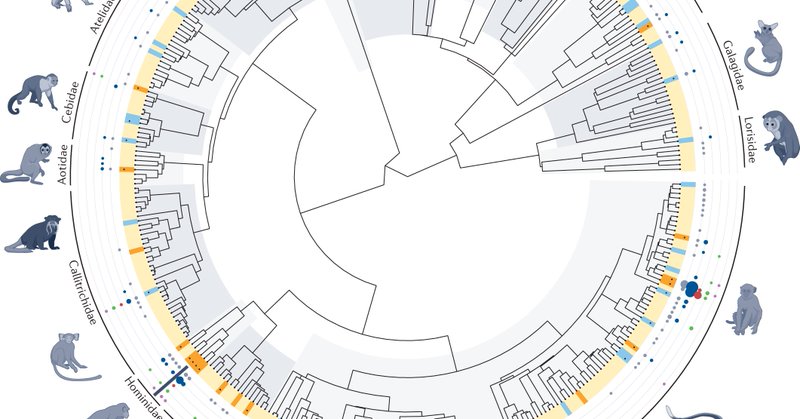
Primate genomics: novel approaches for understanding evolution and disease https://t.co/bkZGlnM2Pm
#Review by David Juan, Gabriel Santpere, @joannalkelley, @ocornejoPopGen & @tmarquesbonet
nature.com
Nature Reviews Genetics - In this Review, the authors describe how advances in comparative primate genomics — complemented by multi-layered omic resources and primate cell systems — are...
1
37
71
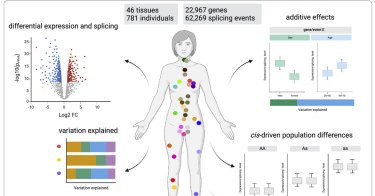
Which individual trait has the largest impact on the transcriptome? Sex, age, genetic ancestry or BMI? Check out our latest publication from the @mele_lab at @CellGenomics to answer this question https://t.co/e8YVpspwm1. Tweetorial 👇🧵
cell.com
García-Pérez et al. perform a multi-tissue analysis of the association between demographic and clinical traits and human transcriptome variation. Ancestry, sex, age, and BMI and certain diseases make...
3
51
136
Our paper on isoform evolution in primates is out in @genomeresearch. Congratulations to the team leading the effort @IBE_Barcelona, @l_ferrandez, Xiaoyu Zhan, @tmarquesbonet, @david_de_juan and Guojie Zhang @TheBgiGroup and many others! https://t.co/TzU6At0Zlj: (1/10).
3
48
131
Avui surt publicat l’article del darrer capítol de la meva tesi doctoral. Un estudi sobre poblacions salvatges de ximpanzé a través de mostres no invasives. Hem descrit amb molt detall la seva història demogràfica i les connexions entre comunitats tan recents com històriques.👇🏻🧵
After 6 very long years, I am very pleased to share with the community our paper entitled: “Population dynamics and genetic connectivity in recent chimpanzee history”. Last chapter of my PhD thesis is out (1/13) #noninvasive #chimpanzee #genomics #popgen
https://t.co/X6xoEB3tpq
1
2
13
📢WE ARE HIRING!! 1 new Postdoc position to join @MarquesBonetLab lab 🧬🙈🙉🙊 at @IBE_Barcelona led by @tmarquesbonet. Starting before fall 2021. Check out and #joinourteam! https://t.co/egcUKxT6G1
#IBEjoboffer
#discoveringlife
#unravelingevolution
#preservingbiodiversity
0
15
15
Out now: HuConTest - testing human contamination in great ape samples. https://t.co/0K8PtJ6IkU with @ClaudiaFontsere @Sojung_B @mae_minaa @tmarquesbonet @MarquesBonetLab
academic.oup.com
Abstract. Modern human contamination is a common problem in ancient DNA studies. We provide evidence that this issue is also present in studies in great ap
0
8
20
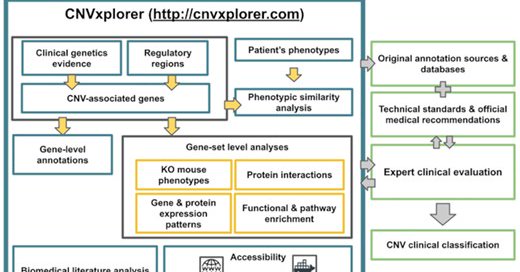
Our open-source webserver for the clinical assessment of CNVs in rare disease patients is now out in @NAR_Open
https://t.co/PIDKPDsvMr Give it a try at https://t.co/unnIEHLK4K Great job of @datarequena @InstitutImagine and the cytogenetics service @hopital_necker @aphp
academic.oup.com
Abstract. Copy Number Variants (CNVs) are an important cause of rare diseases. Array-based Comparative Genomic Hybridization tests yield a ∼12% diagnostic
0
7
14
@MarquesBonetLab A project that started almost 5 years ago. I am particularly proud to have started discussing it with JL Gomez Skarmeta. We dedicate this manuscript to the memory of Jose Luis Gomez Skarmeta. A leader, an inspiration and a friend. @CABD_UPO_CSIC
0
2
12
It has been a privilege to witness how you (@RaquelGarcaPrez and @PaulaEsteller) have made such outstanding work!! Thank you so much for your dedication and patience, to @tmarquesbonet and @MarquesBonetLab for their support!
0
2
11
We are happy to see our latest work now published in @NatureComm. A comprehensive characterization of the epigenetic landscape of LCLs in primate species. @RaquelGarcaPrez @PaulaEsteller @david_de_juan @tmarquesbonet (1/4). Image by @mae_minaa.
3
34
99
Many thanks to the amazing Aitor Serrer-Armero for terrific and long-term work, to @tmarquesbonet for resourceful advice and continuous support in a difficult situation, and to Elaine Ostrander(@NHGRI), @Brian_W_Davis, and @JocelynPlassais for their essential contribution! 6/
0
0
3
9 trait associations with CNVs are involved in chromatin interactions. Among them, we found an interaction between a CNV near MED13L and TBX3 associated with dog height. Also, the interaction of a CNV in an unannotated region with MAP2K6 is associated with hair length. 5/
1
0
1
41 CNV loci were associated with breed differences in certain disease propensities (cataract, retinal atrophy, elbow and hip dysplasias, patellar luxation, …). Interestingly, a higher CNV in DMBT1 was associated with a lower propensity to suffer retinal atrophy. 4/
1
0
0
Several trait associations are enriched in CNVs with lncRNAs expressed in concordant tissues (intelligence with brain, temperament with adrenal gland, racing with blood, muscle and heart), suggesting a role of lncRNAs in these phenotypes that deserves further investigation. 3/
1
0
0
We generated CNV maps for +300 dogs of +200 breeds and identified 96 loci with CNV differences associated with breed morphometric and disease susceptibilities. Integrating these results with chromatin interactions, lncRNA expression and SNVs points to promising loci and genes. 2/
1
0
0
Excited to share our latest work https://t.co/IQ1fhM2Tdl @genomeresearch. The first comprehensive CNV-based GWAS in dogs for breed-specific phenotypes. Amazing collaboration @MarquesBonetLab+Elaine Ostrander(@NHGRI)+@Brian_W_Davis with a fantastic work of Aitor Serres-Armero! 1/
1
8
21
So excited about this! A deep dive into #comorbidities by @jonsv89 & collabs @BSC_CNS @CNIO_Cancer @IBE_Barcelona @INB_Official @CIBER_SAM @univamu @crctoncopole spotting patient subgroups, molecular bases and impact of drugs on disease risk @NatureComms
https://t.co/0L0hJgYU8q
1
6
18
Excellent to see this finally published, a story of perseverance by @jonsv89, my first co-supervised PhD student! Interesting and not entirely expected results that we hope will establish the importance of patient characteristics in studying comorbidities.
A patient gene expression similarity approach leads to new perspectives in disease comorbidity. The work,led by BSC, goes one step further by investigating comorbidities at the molecular level for hundreds of diseases and thousands of patients https://t.co/mdO6ujHgys
@NatureComms
5
8
36
Don't miss the latest #HeredityPodcast featuring @ClaudiaFontsere and @FrandsenPtr where they discuss their most recent work on chimpanzee conservation!
New #HeredityPodcast: Targeted conservation of endangered chimps @FrandsenPtr & @ClaudiaFontsere discuss a new molecular approach to conservation Listen now https://t.co/aZk3tKiShN Read the study https://t.co/Dx4mISRThb
@CopenhagenZoo @MarquesBonetLab @IBE_Barcelona @GenSocUK
0
9
11