Camilo Parada 🦋 @cahuparo.bsky.socia
@cahuparo
Followers
1K
Following
5K
Media
113
Statuses
771
Postdoctoral Scholar at Grünwald and Chang Labs | #OregonState | Plant Pathologist | Colombian | Antiracist | he•his | #FFARfellows | all opinions are my own
Philomath, OR
Joined July 2014
@CornellCALS @LinaQuesadaO I have also received so much support and advice from my peers at NC State 🐺 and Oregon State University 🦫. Thank you all. I can’t wait to get started this September in Ithaca. ¡Nos vemos pronto!.
1
0
8
@CornellCALS This would not have been possible without the deep, long-term support of my mentors. @LinaQuesadaO, Nik Grunwald , Jeff Chang, Peter Balint-Kurti, and Rick Davis.
2
1
7
@CornellCALS It is hard to put into words what this moment means. I am stepping into a dream role where I get to support growers, advance plant health, mentor the next generation of translational plant pathologists, and contribute to NY agriculture through research and extension.
1
0
5
🚨 Big news! I am thrilled to share that I’ll be joining @CornellCALS this fall as a tenure-track Assistant Professor of Field Crop Pathology in the School of Integrative Plant Science! 🌽🫛🌾🐮 #NewPI #PlantPathology #LandGrantMission #Cornell #Extension #CornellSIPS.
13
6
70
RT @maxewilkinson: My favourite discovery ever has just come online. Can I please tell you about some seriously wacky molecular biology? Th….
0
1K
0
RT @QuesadaLabNCSU: @EurekAlert @aaas @plantdisease did a great feature on @cahuparo's sweetpotato-C.fimbriata paper in @MPMIjournal! Check….
eurekalert.org
Sweetpotato black rot is a devastating disease caused by the fungus Ceratocystis fimbriata. In 2015, all sweetpotato-producing states in the United States experienced one of the worst outbreaks...
0
4
0
Excited to see our research on sweetpotato black rot featured! This work from my PhD thesis identifies key effector proteins in Ceratocystis fimbriata, offering targets for functional characterization. Thanks to @QuesadaLabNCSU for your support! #PlantPathology @Sweet_Taters.
@cahuparo, @mnstahr, Childs, and @LinaQuesadaO identify the effector repertoire of the sweet potato fungal pathogen Ceratocystis fimbriata. The expression profiles of these effectors were further evaluated during infection. Find out more here:
3
7
41
RT @CristianOlayaA: Colombian phytopathologists at @plantdisease Pacific Division 2024 @Johadelcpp @cahuparo
0
1
0
RT @mijangos_luis: 🚀 Excited to share our #PopGenInR workshop resources with everyone! 🌍 Uniting for biodiversity with genetics & R. Dive i….
0
120
0
RT @GeneticLiteracy: France's organic sector faces a crisis, with 2022 seeing a 40% drop in areas starting organic conversion and farmers'….
geneticliteracyproject.org
The 21st Agence Bio barometer sheds new light on the causes of France's disenchantment with organic products.
0
14
0
RT @NaturePlants: New Review: "Technology-enabled great leap in deciphering plant genomes" Analysis of 3,517 de no….
0
97
0
RT @JournalSpectrum: Phylogenomics of 710 fungal genomes from the biomedically and technologically important genus Aspergillus reveal that….
0
25
0
RT @lh3lh3: Minimap2-2.27 released with new presets for accurate Nanopore reads and Illumina Complete Long Reads (both recommended by vendo….
github.com
Notable changes to minimap2: New feature: added the lr:hq preset for accurate long reads at ~1% error rate. This was suggested by Oxford Nanopore developers (#1127). Whether this preset works wel...
0
99
0
RT @razoralign: Chrom-pro: A User-Friendly Toolkit for De-novo Chromosome Assembly and Genomic Analysis https://t.c….
0
54
0
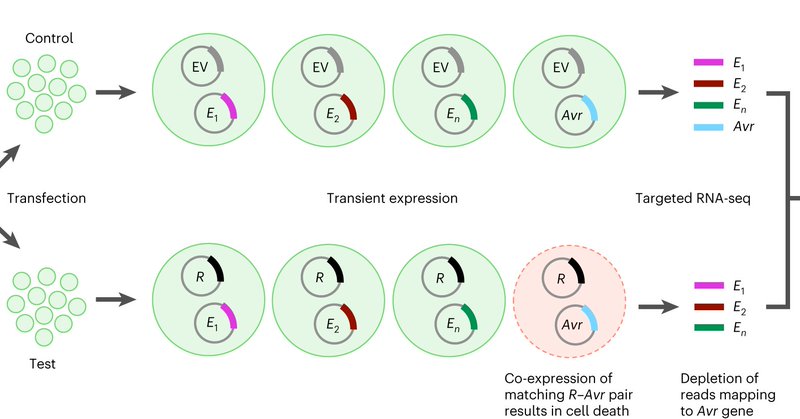
RT @JSperschneider: Out now!🔥Our platform to rapidly screen a large effector library in a single protoplast transformation. Screening of 69….
nature.com
Nature Plants - The authors developed a platform for rapid identification of interacting plant immune receptors and pathogen avirulence proteins by library screening in protoplasts, then used it to...
0
33
0
RT @yuanqinoffice: A new tool is online~.shinyTempSignal: an R shiny application for exploring temporal and other phylogenetic signals .#Mo….
github.com
Explore Temporal and Other Phylogenetic Signals. Contribute to YuLab-SMU/shinyTempSignal development by creating an account on GitHub.
0
6
0
RT @strnr: Haplotype-aware sequence alignment to pangenome graphs 🧬🖥️
github.com
Long-read aligner to pangenome graphs. Contribute to at-cg/minichain development by creating an account on GitHub.
0
11
0
RT @PlantoPhagy: Like this paper a lot: Worth reading just for this sentence: "They also illustrate how the study o….
0
2
0