Adam Weiner
@adamcweiner
Followers
216
Following
614
Media
18
Statuses
89
Postdoc w/ @BeEngelhardt lab at @GladstoneInst | PhD w/ @SohrabShah lab at @CompOncMSK | @UCLA & @aarmey lab alum | ML-omics to study cancer and immunology
San Francisco, CA
Joined September 2019
I officially started my postdoc in @BeEngelhardt 's lab at @GladstoneInst last month!! I’m working with some phenomenal collaborators to improve cancer immunotherapy using genomic and spatiotemporal data. Excited to meet those at the @UCSF Mission Bay and @Stanford campuses!
1
1
26
Thanks to all my co-authors including @marcjwills_, Hongyu Shi, @ivazquezgarcia, @GreedyApple, @SohrabShah, Andrew McPherson, and the rest of the Shah Lab. I learned so much from all of you along the way. 10/.
1
0
1
While most of the paper is similar to the preprint (thread here . I want to draw attention to two new results in Figs 5 & 6 that matured during the peer review process. 3/.
Interested in DNA replication and genomic instability? Our new method PERT identifies S-phase cells and estimates replication timing from single cell whole genome sequencing (scWGS) data with heterogeneous copy number 1/10.
1
0
0
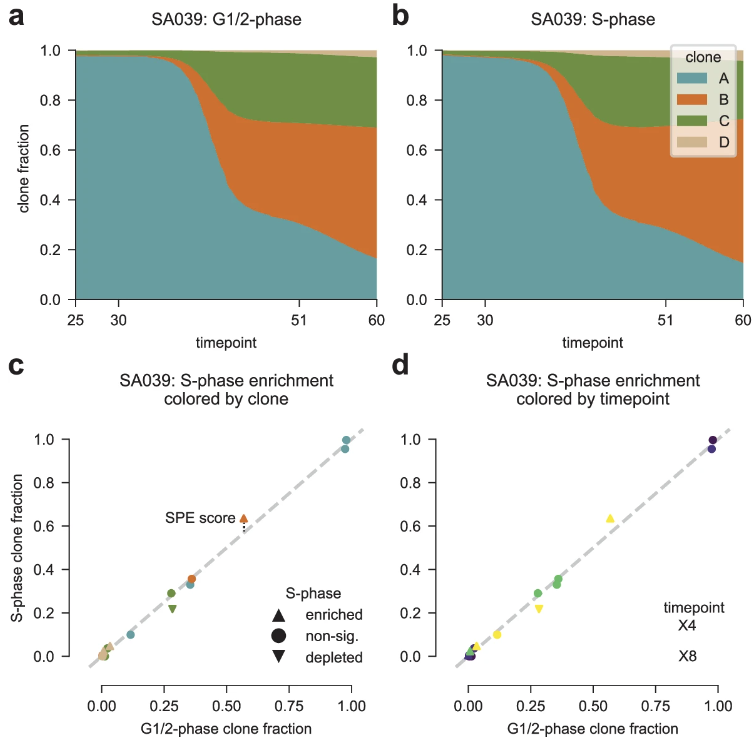
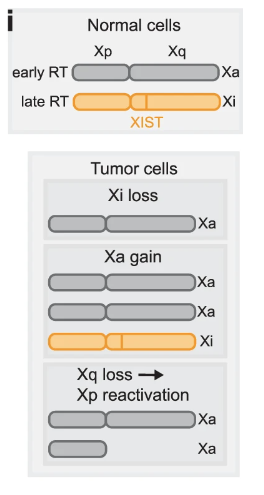
Happy to announce that PERT is now published in @NatureComms ! With PERT, we explore replication timing variability (Fig 3-4), chrXi reactivation (Fig 5), and how S-phase fractions struggle to approximate clone fitness (Fig 6). 1/n.
2
17
44
RT @MaryamPourm: Very excited to share our article detailing spatial features of Epstein-Barr virus positive and negative classic Hodgkin l….
0
4
0
RT @minsoo_kim1: Thrilled to announce that my PhD thesis work has been published in Nature Genetics! 🎉 Huge thanks to my advisors, @SohrabS….
0
5
0
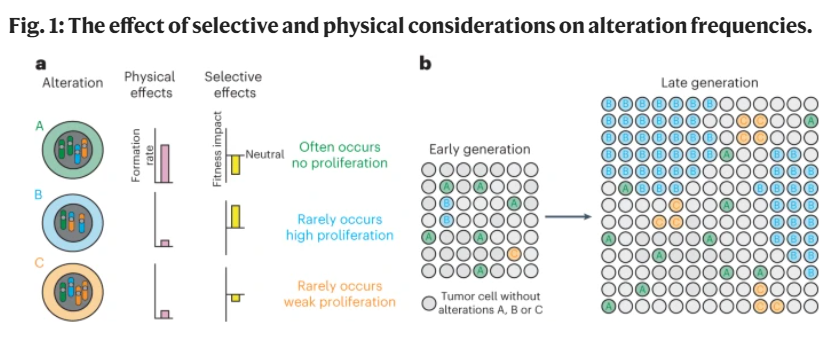
This is one of the better reviews I've read. We have to think about both formation rates and fitness impacts to understand how complex events contribute to tumorigenesis, mets, drug resistance, etc. Worth the read even if you've been thinking about these problems for a while.
NEW REVIEW ARTICLE @NatureCancer – “Aneuploidy and complex genomic rearrangements in cancer evolution” by @VanLooLab #TobyMBaker #SaraWaise #MaximeTarabichi. Read it here👇.
0
0
10
RT @MildredUnti: So excited to announce that my most recent work has been published. If you’re interested in genetically encoding circular….
0
11
0
RT @mmdarmofal: Treatment options are limited for the 3-5% of patients who have cancers of unknown primary. So, we are excited to present 💥….
0
18
0