Wouter Saelens
@Zouters
Followers
189
Following
997
Media
3
Statuses
98
Post-doc at Bart Deplancke lab @ EPFL
Lausanne, Switzerland
Joined March 2016
Still, there's much left to explore, as we "just" implemented two models here. We're working on integrating the sequence directly, and also thinking about interfacing with time dynamics tools.
biorxiv.org
Machine learning methods that fully exploit the dual modality of single-cell RNA+ATAC-seq techniques are still lacking. Here, we developed ChromatinHD, a pair of models that uses the raw accessibil...
0
0
0
ChromatinHD is available at . It's still in beta, but you can already run the main models and interpretation tools. The full-featured package, interfacing with e.g. SCENIC+ @steinaerts, is coming soon. We're happy to hear any feedback.
github.com
High-definition modeling of chromatin + transcriptomics data - DeplanckeLab/ChromatinHD
1
0
0
Ever looked at your ATAC-seq signal and thought: there must be a better way to model this than peaks or windows? Happy to present ChromatinHD, a method that models scATAC+RNA data using the raw fragments. With @BartDeplancke @OlgaPushkarev
biorxiv.org
Machine learning methods that fully exploit the dual modality of single-cell RNA+ATAC-seq techniques are still lacking. Here, we developed ChromatinHD, a pair of models that uses the raw accessibil...
4
33
126
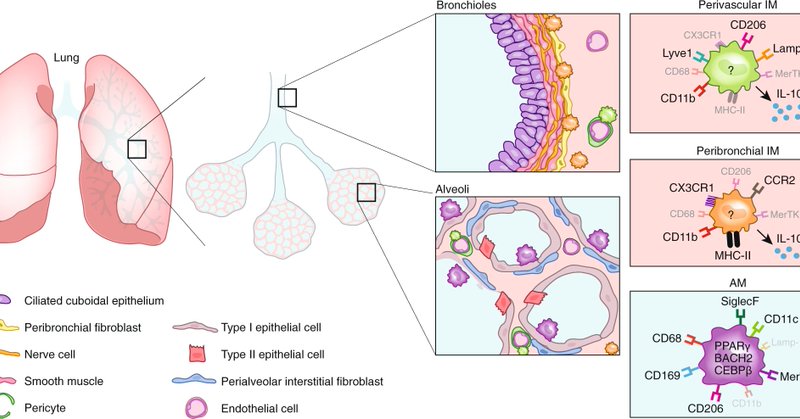
RT @FreyaRSvedberg: The @NatImmunol perspective @MartinGuilliams and I wrote is out. We ask does tissue imprinting restrict macrophage plas….
nature.com
Nature Immunology - Based on the results of recent studies that have dissected the response of individual macrophage subsets to pulmonary insults, Guilliams and Svedberg call for an adjustment of...
0
66
0
RT @NatureBiotech: Generalizing RNA velocity to transient cell states through dynamical modeling .
0
127
0
RT @cziscience: #Opensource software is essential to science. That’s why we’re awarding 23 grants to support open source tools that acceler….
chanzuckerberg.com
CZI supports software projects that accelerate biomedical research
0
98
0
RT @danwagnerlab: With widespread adoption of single-cell-omics technologies, developmental biologists are increasingly relying on the use….
0
165
0
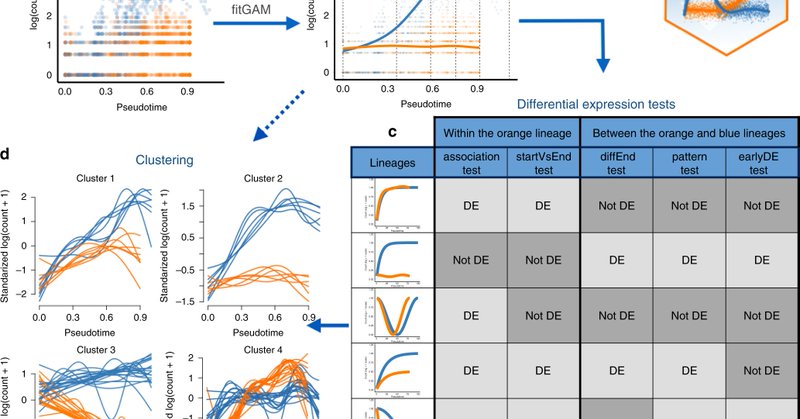
RT @koenvdberge_Be: Our work on differential expression (DE) analysis downstream of trajectory inference (TI) is now published! A small thr….
nature.com
Nature Communications - Downstream of trajectory inference for cell lineages based on scRNA-seq data, differential expression analysis yields insight into biological processes. Here, Van den Berge...
0
39
0
RT @fabian_theis: We're excited to release a #scanpy update for the analysis and visualization of #spatialTranscriptomics data, focussing o….
0
107
0