Yoseph Barash
@YosephBarash
Followers
957
Following
4K
Media
158
Statuses
2K
@[email protected] Opinions - own. Lab @PennGenetics , @CIS_Penn , @UPennIBI
Philadelphia, PA
Joined July 2015
RT @pkoo562: Our work on "Evaluating the representational power of pre-trained DNA language models for regulatory genomics" led by @AmberZq….
genomebiology.biomedcentral.com
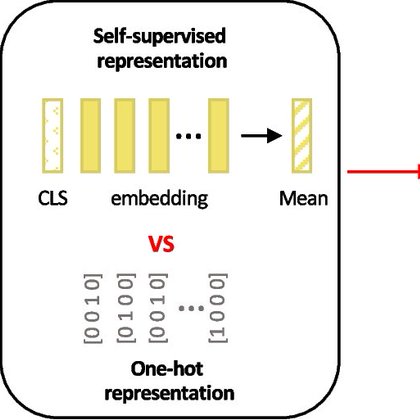
Background The emergence of genomic language models (gLMs) offers an unsupervised approach to learning a wide diversity of cis-regulatory patterns in the non-coding genome without requiring labels of...
0
42
0
RT @seemaychou: I worry the pendulum has swung too far towards generalists over specialists, sacrificing wisdom for speed. Resulting in eit….
0
10
0
RT @KipoiZoo: Join us for our next Kipoi Seminar with Yoseph Barash,University of Pennsylvania.@PennGenetics.@CIS_Penn.@UPennIBI.@YosephBar….
0
1
0
Given some recent publications regarding #RNA #Splicing prediction using #DeepLearning we decided to assess some of those claims with surprising findings that have to do more with the target function you chose…
biociphers.wordpress.com
In today’s blog post, I want to discuss some work/claims related to a topic that has been one of my lab’s focus areas – predicting tissue-specific alternative splicing (AS) of RNA. Some backg…
1
17
45
RT @JMTali: Excited to share our latest work! We asked if and how TDP-43 regulates RNA transport in neurons and if this is misregulated in….
0
45
0
RT @zhangf: We are excited to report the discovery of TIGRs, a widely-occurring RNA-guided system found in bacteria and their viruses. TIGR….
0
264
0
RT @AntonIPetrov: 🚀 New Paper Alert! 🚀. 𝐑𝟐𝐃𝐓: a comprehensive platform for visualising RNA secondary structure. Check it out in @NAR_Open:….
0
14
0
@SashaGusevPosts @jkpritch @mstephens999 @tuuliel_lab @hjpimentel @kauralasoo As we also wrote in the blog post - This work started many years ago with the late and great @casey6r0wn who kept giving us feedback/insights. We miss him as a friend and colleague. We dedicated the work to Casey and his contributions to Science 💔.
0
0
2
We much appreciate constructive feedback on the ideas/analysis from ppl in this area e.g. (please ignore if you don't have the bandwidth/interest or forward to others I may have missed. ) - Tx! @SashaGusevPosts @jkpritch @mstephens999 @tuuliel_lab @hjpimentel @kauralasoo.
1
1
1
📣 Our 2nd Blog post on improving #sQTL modeling and why it matters #RNASplicing #Genetics #Statistics #QTL
biociphers.wordpress.com
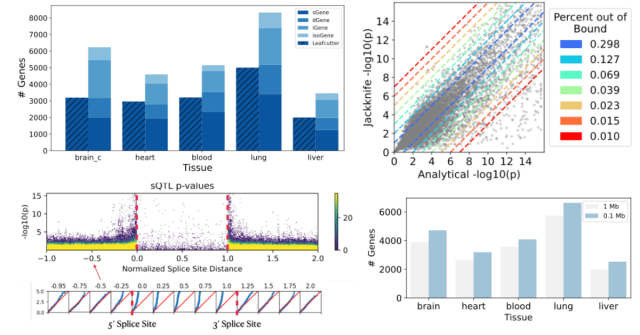
In my previous post, I described how, in our recent sQTL preprint, we broke current sQTL pipelines into their basic elements. This led to identifying potential issues/areas we may be able to improv…
1
2
8
📣 New post re our recent findings/methods dev for #sQTL #RNA #Splicing #RNA-Seq. We think this will be highly useful/impactful for (s)QTL detection/quantification. Hope to get (constructive 😉) feedback and much usage from the community! 😀
biociphers.wordpress.com
The next few blog posts will be dedicated to a high level overview of our preprint on splicing QTL (sQTL) modeling: “A Deep Dive into Statistical Modeling of RNA Splicing QTLs Reveals New Variants …
0
5
19
RT @andrei_thomas_t: An #RNA #splicing project 3 yrs in the making finally sees the light of day. Microexons, meet macroglia… cancerous mac….
biorxiv.org
To overcome the paucity of known tumor-specific surface antigens in pediatric high-grade glioma (pHGG), we contrasted splicing patterns in pHGGs and normal brain samples. Among alternative splicing...
0
5
0
RT @shramdas: Our mentor Casey Brown, apart from being an extraordinary scientist, was a brilliant teacher who loved everything genetics. I….
youtube.com
0
27
0
RT @pkoo562: I'm hiring:. 1. PhD-level Research associate to generate perturbation data. 2. ML postdoc to build multimodal generative AI fo….
0
37
0
RT @gxr: Where RNA Science Meets AI, May 4–8, 2025, Ascona. Invited speakers: @OliverStegle @fabian_theis @fiddle @EvaMariaNovoa @BTreutlei….
0
9
0
RT @AI4Biology: G4mer: RNA language model identifies G-quadruplexes and disease variants across transcriptome. AI analyses population-scale….
biorxiv.org
RNA G-quadruplexes (rG4s) are key regulatory elements in gene expression, yet the effects of genetic variants on rG4 formation remain underexplored. Here, we introduce G4mer, an RNA language model...
0
4
0
RT @quaidmorris: 💥💥💥New preprint 💥💥💥on the EuPRI resource. Where we reconstruct the motifs for 34,736 RBPs and study their function and evo….
0
16
0
RT @andrei_thomas_t: Yet more ways alt #RNA #splicing drives #cancer chemoresistance. Take-home messages: a) some drugs targeting purine me….
aacrjournals.org
Alternative splicing is a potent mechanism of acquired drug resistance in relapsed/refractory acute lymphoblastic leukemias that has diagnostic and therapeutic implications for patients who lack...
0
4
0
RT @pkoo562: Are you a wet lab biologist ready to shape the future of AI in genomics? CSHL is hiring a PhD-level Research Associate to join….
0
27
0