WESTPA Software
@WestpaSoftware
Followers
256
Following
26
Media
4
Statuses
56
Moved @westpasoftware.bsky.social. Highly scalable software for weighted ensemble simulations with any dynamics engine, including MD and systems biology.
Joined January 2019
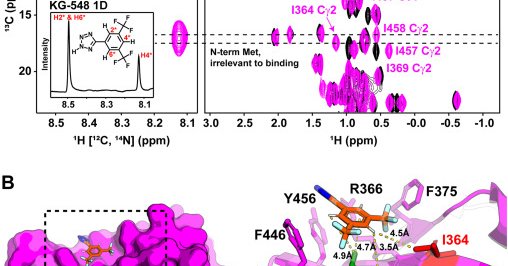
The bulk of my thesis work was recently published! We used 19F NMR and weighted ensemble simulations among other methods to explore hidden dimer states of the HIV-1 capsid protein. If this sounds interesting to you, see the full paper here: https://t.co/beUckC2RTl
1
3
5
Just out in @JChemPhys! Integrating Weighted Ensemble (@WestpaSoftware) with OPES-Flooding for accurate and efficient calculation of rare-event kinetics involving barrier crossing and diffusion (e.g. protein folding, ligand binding) @uoCHandBIC #compchem
https://t.co/5ynY6NANbi
0
22
83
Check out this recent publication from the @agrossfield lab where they used WESTPA to determine the effects of system size on free energy landscapes of a phase-separating ternary lipid bilayer! https://t.co/dg6aL0avBz
pubs.aip.org
The “lipid raft” hypothesis proposes that cell membranes contain distinct domains of varying lipid compositions, where “rafts” of ordered lipids and cholesterol
0
2
13
New work on protein design... 🚨S2 immunogen design using advanced simulation @WestpaSoftware WE MD--> opening mechanism--> design--> stabilized protein--> cryoEM structure + immunology @xandra_nuqui @LCasalino88 @McLellan_Lab @ChandranLab
https://t.co/Nhm580Tsom
2
38
198
We are excited to share this collaboration between the Gardner and Chong labs in which weighted ensemble simulations are combined with NMR and ensemble docking!
jbc.org
Transcription factors are challenging to target with small-molecule inhibitors due to their structural plasticity and lack of catalytic sites. Notable exceptions include naturally ligand-regulated...
1
7
25
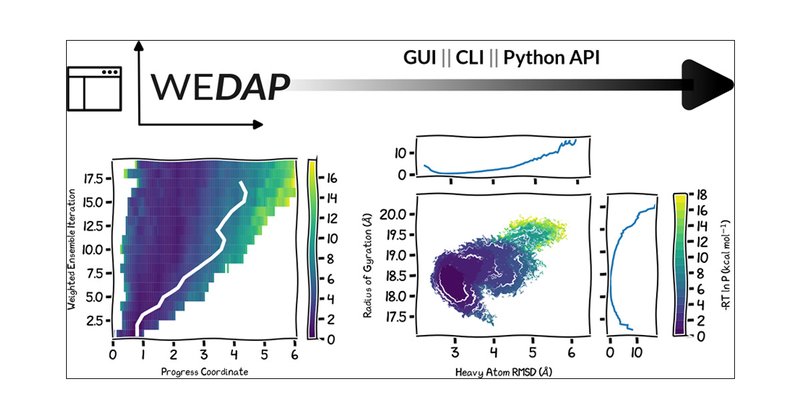
#HighlightOfTheWeek Animated time-evolution of a 2D probability distribution obtained using WEDAP, a python package to plot molecular simulation data. Work by @darianyang and @ltchong. #compchem
https://t.co/AFOKlY9pXr
1
15
39
We're excited to see Shirley Ahn's group using WESTPA to explore optimizing weighted ensemble simulations with cumulative distribution functions!
pubs.aip.org
Molecular dynamics (MD) simulations are powerful tools for studying the dynamic behavior of molecular systems at the atomic level. MD simulations provide insigh
0
7
8
Want to try it out? Just run: "pip install wedap" - PyPI: https://t.co/ZGpkGSoMFH - GitHub: https://t.co/2FYuXUXuGI - Docs Page: https://t.co/hHjPGiIxs2 (4/4)
0
0
1
WEDAP works with both WE and standard MD data through multiple interfaces: a GUI, a CLI, and a Python API (2/4)
1
0
1
Having trouble visualizing your data from WESTPA? Try WEDAP! A user-friendly plotting package from @darianyang and @ltchong (1/4) https://t.co/yofj2KDidD
pubs.acs.org
Given the growing interest in path sampling methods for extending the time scales of molecular dynamics (MD) simulations, there has been great interest in software tools that streamline the generat...
1
13
36
Thrilled to share that my thesis manuscript is out! Here, we used WESTPA2 to show that mutations in Abl1 lead to drug resistance by dramatically shifting relative state populations, impacting the conformational search step of binding. Link below: https://t.co/q1TnDNQfPA
biorxiv.org
The intrinsic dynamics of most proteins are central to their function. Protein tyrosine kinases such as Abl1 undergo significant conformational changes that modulate their activity in response to...
1
4
19
Revisiting Textbook Azide-Clock Reactions: A “Propeller-Crawling” Mechanism Explains Differences in Rates | Journal of the American Chemical Society @ltchong @PittChem @PittTweet #Azide #Clock #PropellerCrawling #Mechanism #Rates
0
6
24
Another great BPS with both vintage and fresh WESTPA developers/users from the @ltchong @RommieAmaro @DMZuckermanLab @Terrasztain and Ahn Labs!
1
3
16
Register now for the free WESTPA Workshop @Pitt_Chemistry, June 20-21. Explore rare event simulations like protein folding, ligand binding, & chemical reactions with our open-source software. Perfect for those using AMBER, NAMD, GROMACS, etc. Reg info: https://t.co/JeQwUMDAIq
docs.google.com
Please join us for this year's WESTPA workshop at the University of Pittsburgh, June 20-21, 2024. We will be celebrating the 10th anniversary of WESTPA! WESTPA is an open-source, highly-scalable...
0
14
39
Using WE simulations and experiments, @ChongLabPitt and the Horne lab explore the atomistic folding pathways of protein mimetics https://t.co/wskDlqwakv
pubs.rsc.org
Sequence-encoded protein folding is a ubiquitous biological process that has been successfully engineered in a range of oligomeric molecules with artificial backbone chemical connectivity. A remark...
0
0
0
Want to see WE / WESTPA applied towards understanding the free-energy landscape of phase separation in lipid bilayers? Check it out:
0
1
4
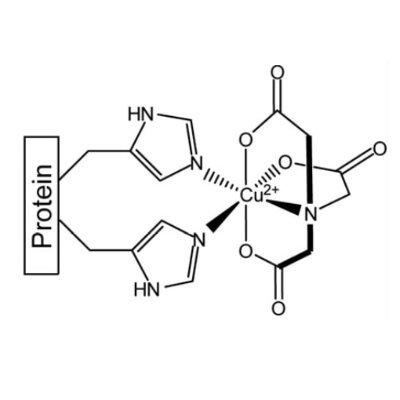
Beyond coarse-grained modeling: Seconds-timescale protein conformational change measured using Cu(II)-based EPR and weighted ensemble MD simulations. @ltchong Congratulations to Xiaowei and @AnthonyBogetti on this work, now published in Protein Science: https://t.co/EE8B2VW8SV
1
12
39
Congrats to one of our core WESTPA developers Jeremy Leung for receiving a MolSSI software fellowship!
We loved having our 2023-24 MolSSI Software Fellows with us last week! We thank Jessica Nash @jessica_a_nash and the other MolSSI Software Scientists for another inspired week of programming and mentorship!
0
0
7
Brossard and Corcelli just published their first paper using WESTPA! They investigated the mechanism for binding a small molecule to the minor groove of DNA. https://t.co/MhT4VIsi3N
pubs.acs.org
Although DNA–ligand binding is pervasive in biology, little is known about molecular-level binding mechanisms. Using all-atom, explicit-solvent molecular dynamics simulations in conjunction with...
0
1
3