VingronLab
@VingronLab
Followers
653
Following
126
Media
16
Statuses
271
regulatory sequence motifs, epigenetic regulation, sequence analysis, single cell transcriptomics, haplotyping, 3D chromatin structure, biological networks
Berlin, Germany
Joined January 2019
We are excited to share 𝘀𝗰𝗔𝗧𝗔𝗰𝗮𝘁, a great new tool for annotating #scATACseq data! .Check out the thread for more details ⬇️.
I am happy to share that my PhD work, 𝘀𝗰𝗔𝗧𝗔𝗰𝗮𝘁🐈, is now out in #nargab @VingronLab.#scATAcat #epigenomics #singlecell.Here is a 🧵👇.
0
0
6
You can try out CAbiNet by simply installing it from our GitHub:
github.com
Contribute to VingronLab/CAbiNet development by creating an account on GitHub.
0
0
0
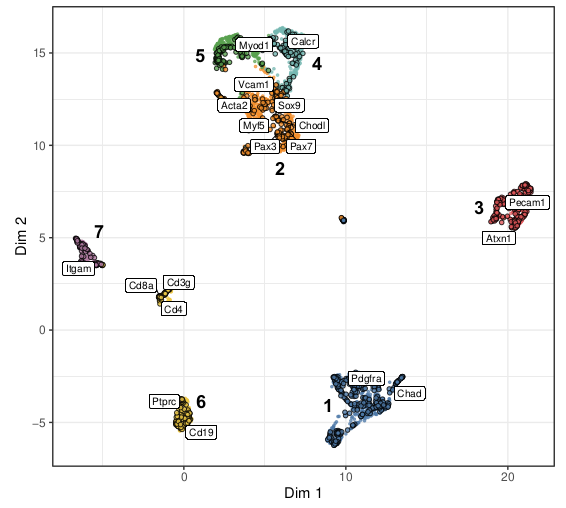
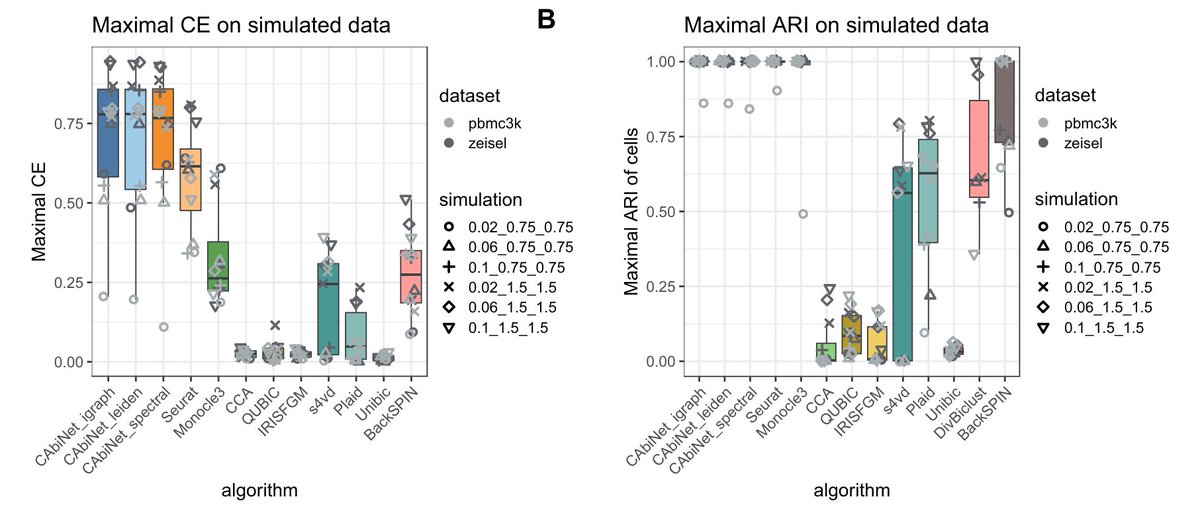
Excited to share our new paper “CAbiNet: joint clustering and visualization of cells and genes for single-cell transcriptomics” and accompanying R package by @young_Yanie_Z and @ClemensKohl which was just published in @NAR_Open !.
academic.oup.com
Abstract. A fundamental analysis task for single-cell transcriptomics data is clustering with subsequent visualization of cell clusters. The genes responsi
1
2
7
Our collaboration paper with @fubcp and @MundlosLab is out! Congrats to all the authors 🥳 Learn more 👇🏼.
I'm very happy to share that our collaborative study between @fubcp and @StefanMundlos & @vingron is finally published online @iScience_CP. We show the mechano-dependency of BMP9 SMAD-signaling in the vasculature. #BMP #ATACseq #genomics #vasculature.
0
0
9
Association plots paper is out! Congrats to @ela_gralinska 🥳 Read the 🧵to learn more 🤓.
**Association Plots** and cluster-specific genes in high-dimensional data. (1/7) Very happy to share that our manuscript on Association Plots from @VingronLab (@MPI_MolGen) , summarizing the basic concepts of my PhD project, is finally out. Below the key points from our paper:
0
1
7
Join us for today’s Institute Social Hour - Easter edition! Egg themed games, chocolates and homemade pizza is waiting for you! @MPI_MolGen
1
1
12
Our new study “Prediction of protein-protein interactions using sequences of intrinsically disordered regions” published in Proteins journal, reveals that intrinsically disordered regions can aid in predicting specific features of eukaryotic PPI network! Check out our paper!.
(1/4).I am happy to share that my PhD work on predicting protein-protein interactions using intrinsically disordered regions is now published in Proteins : Structure, Function, and Bioinformatics ! @VingronLab .#PPIs #IntrinsicallyDisorderedRegions #IDPs.
0
1
19
📢 #JobAlert: Wir haben eine Postdoc Stelle in Bioinformatik an der Freien Universität zu besetzen! .@FU_Berlin @MPI_MolGen.📅 Bewerben bis 20.03.2023!.Bitte verbreiten Sie die Nachricht:.
0
7
9
RT @MPI_MolGen: How can anyone survive a shattered genome? Our @VingronLab @MundlosLab @MeloUira & R. Schöpflin found that the answer lies….
molgen.mpg.de
Scientists have reconstructed the chromosomes of patients with an extremely high number of aberrations in their genome that could alter the expression of nearby genes and potentially cause disease....
0
4
0
Our new work 'CAbiNet: Joint visualization of cells and genes based on a gene-cell graph' by Yan Zhao @young_Yanie_Z, Clemens Kohl @ClemensKohl, Martin Vingron @VingronLab, et. al. has been released at bioRxiv ( ).Looking forward to your comments!.
biorxiv.org
In routine single-cell RNA-sequencing (scRNA-seq) analysis workflows, cells are commonly visualized in 2D to show the patterns in the data. However, these visualization approaches do not give any...
0
5
13
RT @biorxivpreprint: CAbiNet: Joint visualization of cells and genes based on a gene-cell graph #bioRxiv.
0
2
0
We Put Up Our Christmas Tree 🎄 🎅 🤶 Thanks to all Vingron lab members for their collaborative efforts! Come and visit the Pikachu on top of tree 🎊 @MPI_MolGen
0
3
22
Our new #bioconductor R package 'APL' by @ela_gralinska, @ClemensKohl, and Martin Vingron @VingronLab is out!. 'APL' implements the concept of Association Plots for #SingleCell data.
bioconductor.org
APL is a package developed for computation of Association Plots (AP), a method for visualization and analysis of single cell transcriptomics data. The main focus of APL is the identification of genes...
0
6
16
(8/8).Interested? Check out our paper and the #bioconductor R package implementing this concept! . Paper: R package: Questions? @ela_gralinska.
bioconductor.org
APL is a package developed for computation of Association Plots (AP), a method for visualization and analysis of single cell transcriptomics data. The main focus of APL is the identification of genes...
1
2
5