Usman Enam
@UsmanEnam
Followers
265
Following
315
Media
7
Statuses
117
Looking for weird bases in weird places. Genetics PhD student in the Fire Lab @StanfordBiosci Cover photo: work of @dsgoodsell
Joined March 2012
Out now in Applied and Environmental Microbiology! We developed a super fast and super easy approach to enrich for genomes of interest in metagenomic samples based on differential modification status✂️🧬: https://t.co/eYuDoieJH4 1/9
journals.asm.org
Pathogens that contaminate the food supply or spread through other means can cause outbreaks that bring devastating repercussions to the health of a populace. Investigations to trace the source of...
3
6
32
My article in today’s Dawn depicts the state of CP in 🇵🇰. We @CCR_pk have written an Integrated Child Protection System & urge the fed & prov govts to adopt it to effectively manage CP in Pak. @saynotoweapons
@rabiaazfar
@shaiyannemalik
@ZubiaMotiwalla
https://t.co/jfDsPbGt7j
dawn.com
Each new tragedy eclipses the previous one in the public’s memory.
2
4
11
I’m amped to share our new work to decrypt transcriptional repressors, now on bioRxiv: https://t.co/BeKDmg3rUy We paired deep mutational scans with machine learning to predict and even tune repressor activity. (1/8)
4
27
94
STARTING A NEW COMPANY IS A CREATIVE PROCESS. Every detail, even the name, is part of this journey. As with all creative endeavors, it’s crucial to be selective about whose feedback you consider. Accepting feedback from everyone will strip away the uniqueness critical for
16
15
130
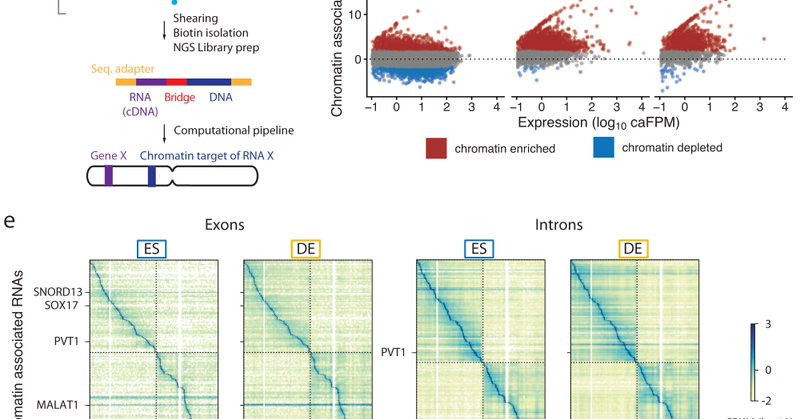
It's out! Our paper describing chromatin interacting RNAs during human ES cell differentiation just published https://t.co/AlB84QXSBw with @climouse @SmithOwenK @fryer_kelsey @dmjukam @WJGreenleaf rethinking chromatin-RNA contacts in gene expression and genome organization
nature.com
Nature Communications - Many types of RNAs are associated with chromatin. Here the authors identify chromatin-bound RNAs and their binding sites in human embryonic stem cells suggesting that most...
6
25
115
Incoming first-year graduate students, especially those from URM backgrounds - check out our GSEC mentorship program! Applications due by THIS SATURDAY (5/20) for both mentees and mentors - info & apply below ⬇ https://t.co/TUfjdY448r
https://t.co/Pm5LITsu8r
cientificolatino.com
Científico Latino Graduate Student Engagement and Community (GSEC) Program focuses supporting GSMI scholars and all diverse gradstudents as they navigate their first year in graduate school.
Are you about to start a STEM grad program?🧪 Científico Latino’s GSEC program (Grad Student Engagement & Community) is now OPEN to the public! https://t.co/TUfjdY448r 1st year grad students & mentors – apply by May 20! 🧵 1/5
0
28
26
I only sterilize my media in gas autoclaves. Electric autoclaves just don't give me the caramelization I want in my LB
1
0
11
Next up: NCBI BLAST moving to a subscription based model. 5 free searches/month, $20 to access results with E values < 0.005. Premium tier includes blue checks next to refseq alignments
Just got the email from @Quartzy that we will have to pay $159/month for the database functions that have been free for the last 10+ years. I still have the email from Jan 2016 that reads: “What the future holds: Quartzy is and always will be free for scientists” 🙈🤬😭
1
0
12
A lack of restrictions is the greatest enemy of creativity. The architects, painters, and calligraphers who decorated these mosques were limited by religious doctrine - but that forced them to find a route to extraordinary beauty, one they might not otherwise have found.
57
258
3K
My final essay for 2022 🧬 The genomics revolution is now enabling a broad exploration across the Tree of Life. Let's explore the wild world of Extreme Biology:
5
50
339
I also want to point you toward a highly complementary and exciting approach/study from @iamfanggang's group led by @clannabel7 called mEnrich-Seq who also took the effort to assess the broad applicability of an approach like this: https://t.co/UxE4hGmoSS 9/9
0
0
4
This study was done with much discussion and collaboration from @RZheludev, Josh Cherry at the NCBI, Susan Leonard and David Lipman at the FDA, many members of the Fire Lab and Straight Lab and Andy Fire, my PI! 8/9
1
0
3
This is my first paper from my PhD and I am now using REMoDE for an exciting endeavor which is to screen (and enrich) metagenomic samples for genomes with yet undiscovered modifications or non-canonical bases 🧬 7/9
1
0
4
We use the distributive ability of T5 exonuclease to deplete shorter DNA fragments from a reaction before eliminating longer ones. We suggest using this method if you have a lot of samples to process... 6/9
1
0
2
A quick note on size-selection of DNA: While gel electrophoresis and extraction is great at this task, we also developed an alternative method using T5 exonuclease that can be done in the same tube, parallelized, takes <30 mins and costs <3¢ per sample. 5/9
1
0
2
We applied this approach to enrich for E. coli and S. enterica genomes in mock metagenomic samples which are methylated at GATC and CC(A/T)GG sites. We used restriction enzymes that target these sites only if unmethylated. What we observed was up to a 142-fold enrichment!! 4/9
1
0
3
This leads to a clean segregation of size between DNA that is not modified and therefore digested and DNA that is modified and therefore undigested. Size-selecting for high molecular weight DNA then robustly enriches your sequencing dataset for your genome of interest 3/9
1
0
2
The premise of REMoDE (Restriction Endonuclease-based Modification Dependent Enrichment) is simple and relies on enrichment via subtraction. We harness the ability of modified (methylated or otherwise) DNA to resist against the action of specific restriction enzymes 2/9
1
0
3
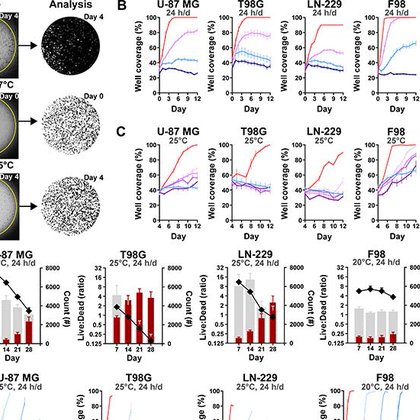
Cooling brain tumors. Prolonging (rodent) lives. 🧠🐭 Out in @ScienceAdvances: https://t.co/Q4K65DZFC2
@dukeengineering @utswnews @AKUGlobal
#glioblastoma #bioengineering #medicaldevices #cancer #brain
science.org
'Cytostatic' hypothermia dampens cellular pathways, safely halts glioblastoma growth in rats, and holds translational promise.
5
15
75
Restriction Endonuclease-based Modification-Dependent Enrichment (REMoDE) of DNA for Metagenomic Sequencing https://t.co/iMcKm5YAb4
#biorxiv_micrbio
0
5
20