Sanjeevi Sivasankar
@Sivasankar_Lab
Followers
244
Following
113
Media
8
Statuses
148
Professor at University of California, Davis. Studying cadherins, desmosomes, biophysics of cell adhesion, molecular biomechanics.
Davis, CA
Joined March 2020
We show that in both epithelial and heart cells, desmoplakin behaves like a 'directional' force sensor, and responds to changes in tensile forces by undergoing a dramatic conformational change.
0
0
1
Desmosomes are adhesive junctions that mediate the integrity of tension prone tissues. Yet, how desmosomes sense and respond to mechanical stimuli are unknown. We show that actomyosin forces alter the conformation of desmoplakin, a key desmosomal protein.
1
0
1
Delighted to announce the publication of our manuscript today in @NatureComms showing how desmosomes sense and respond to mechanical forces. Led by the truly amazing Belle Dong with many thanks to our collaborators in the Choy & Kwiatkowski labs. https://t.co/xgjCPCcGc9
1
0
1
U C Davis has a very nice story on our latest research...
Outside-In Signaling Shows a Route Into Cancer Cells @UCDavisBME @UCDavisCOE
1
1
5
We show that antibodies can trap cadherin in an X-dimer conformation. Formation of an X-dimer triggers the phosphorylation of cytoplasmic p120-catenin, a suppressor of cadherin endocytosis. Consequently p120 dissociates from the X-dimer which causes cadherin endocytosis.
0
0
1
We demonstrate that an extracellular conformation of cadherin, an essential cell-cell adhesion protein, triggers cadherin internalization. Our work provides insights into how cells regulate adhesion and can be exploited to design antibodies for intracellular drug delivery.
1
0
1
Delighted to announce our new publication in @NatureComms on outside-in engineering of cadherin endocytosis using a conformation strengthening antibody. Exceptional effort by co-first authors @BinXie95765695 and Shipeng Xu. Funded by @NIGMS. https://t.co/Y7Vq2MFdum
1
2
4
Sanjeevi Sivasankar and colleagues @ucdavis used MD simulations and single molecule AFM to elucidate the molecular mechanism by which the antibody 66E8 strengthens the interactions of E-cadherin cell-cell adhesion proteins @Sivasankar_Lab
https://t.co/hbLUTcXtIp
0
1
5
Thanks for the invite, Sabyasachi!!
All you need to know to develop and use an Atomic Force Microscope by Sanjeevi Sivasankar...@Sivasankar_Lab
1
0
9
Online now! Strengthening E-cadherin adhesion via antibody-mediated binding stabilization
0
1
6
Our study offers guidelines for designing antibodies that strengthen cadherin adhesion. We show that these antibodies stabilize the primary E-cadherin binding conformation and inhibit conformational changes during dissociation. Strong collaboration with Barry Gumbiner's lab.
0
0
1
Announcing the publication of our paper in @Structure_CP describing the molecular mechanism by which antibodies strengthen the binding of E-cadherin cell-cell adhesion proteins. Fabulous work by Bin Xie and Shipeng Xu @UCD_BPH @UCDavisBME @UCDavisCOE
https://t.co/IYtyqek3PK
1
3
19
This is a devastating loss. Bill’s work in cell adhesion was an inspiration to our lab. And Bill was a most generous and kind colleague. My deepest condolences to his family and friends. RIP.
With sorrow, we (@Stanford_MCP @SSBiophysics) share news that our friend & colleague, Bill Weis, passed away Oct 13,2023. Among many other reasons, he is missed for his discoveries, mentoring, and for the gift of his incisive intellect, wry wit, & kindness. RIP.
1
1
14
@J_Cell_Sci @shafrazomer @ucdavis @UCDavisCOE @UCDavisBME @UCDavis_Egghead has a nice blog post describing this research https://t.co/87g3OOERBi
ucdavis.edu
Biomedical engineers at UC Davis have come up with a new tool for tracing interactions between proteins. The new, light-activated tool could have wide applications in cell biology.
0
0
7
An ingenious technique invented at UC Davis will help biologists track how proteins interact with each other @UCDavisBME @UCDavisCOE @J_Cell_Sci
ucdavis.edu
Biomedical engineers at UC Davis have come up with a new tool for tracing interactions between proteins. The new, light-activated tool could have wide applications in cell biology.
0
6
14
@J_Cell_Sci @shafrazomer @ucdavis @UCDavisCOE @UCDavisBME We benchmark LAB against the widely used TurboID proximity labeling method by measuring the proteome of E-cadherin, an essential cell–cell adhesion protein. LAB can map E-cadherin-binding partners with higher accuracy and significantly fewer false positives than TurboID.
0
0
3
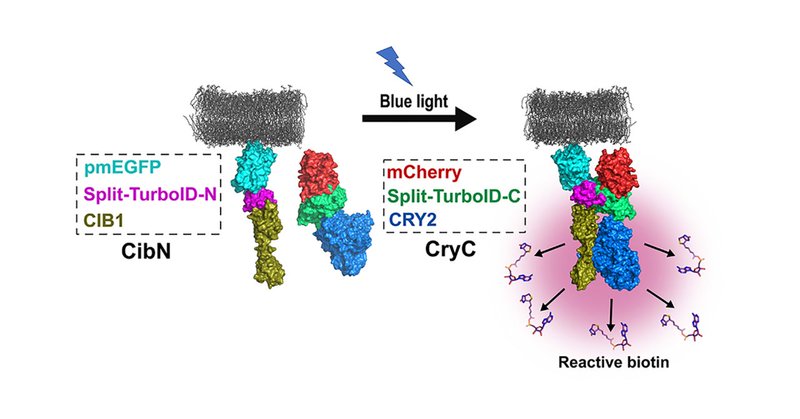
@J_Cell_Sci @shafrazomer @ucdavis @UCDavisCOE @UCDavisBME Our technology, called light-activated BioID (LAB), fuses the two halves of the split-TurboID proximity labeling enzyme to the photodimeric proteins CRY2 and CIB1. Upon illumination with blue light, CRY2 and CIB1 dimerize, reconstitute split-TurboID and initiate biotinylation.
0
0
2
Delighted that our light-activated proximity labeling technology for mapping protein–protein interactions was just published in @J_Cell_Sci Special thanks to co-first authors @shafrazomer and Carolyn Davis @ucdavis @UCDavisCOE @UCDavisBME
https://t.co/2ggPFGMelX
journals.biologists.com
Summary: We present a light-activated proximity labeling technology for mapping protein–protein interactions at the cell membrane with high accuracy and precision.
8
14
71
Engineering the Interactions of Classical #Cadherin Cell–Cell #AdhesionProteins, a #BriefReview by @Sivasankar_Lab @ucdavisvetmed in #JI_BiophysicalImmunology #ReadTheJI 👉 https://t.co/562ja4J8aG
0
2
3
Very cool: Light Activated BioID (LAB): an optically activated proximity labeling system to study protein-protein interactions. | bioRxiv
1
45
242