Roman Barth
@RomanBarth2
Followers
386
Following
311
Media
111
Statuses
323
Postdoc at David Baker lab @ IPD U Washington | PhD at Cees Dekker lab @ TU Delft | Schmidt Science Fellow | Creating and understanding the tiniest machines
Delft, The Netherlands
Joined December 2018
Exciting news 📃🧬🔬 Today we report on the dynamics of a fascinating class of molecular motors, so-called SMC proteins in @CellCellPress
https://t.co/MRIIHE893Z
3
23
116
A huge congratulations to the recipients of the 2025 Weintraub Award! These ten exceptional graduate students exemplify the bold, creative, and pioneering spirit embodied by Dr. Hal Weintraub. https://t.co/DqyAYoQ2ki
0
6
26
Science is a collaborative adventure, and this moment really feels like a celebration of the collective effort, curiosity, and passion that drives research. @TNWTUDelft
1
0
1
I'm excited to share that I've been selected as one of the 10 recipients of the prestigious Weintraub Award from @fredhutch this year! 🏆 I owe a massive debt of gratitude to the remarkable colleagues and mentors who have shaped my path, especially @cees_dekker !!
4
2
18
Check out this cool article by @NewScientistNL on our latest work about direction-switching molecular motors racing along DNA: https://t.co/gvZ0035w0O
0
4
18
Thank you to all that contributed!! Especially to @cees_dekker and the fantastic team at the @IMPvienna Gabriele Litos, Iain Davidson, and Jan-Michael Peters!!
0
0
2
We hope that by disentangling the multiple contributions of the CTCF N-terminus to the stalling of cohesin at CTCF sites, we are another step closer to understanding how cohesin and CTCF control and shape genomes across the tree of life 🌳
1
0
1
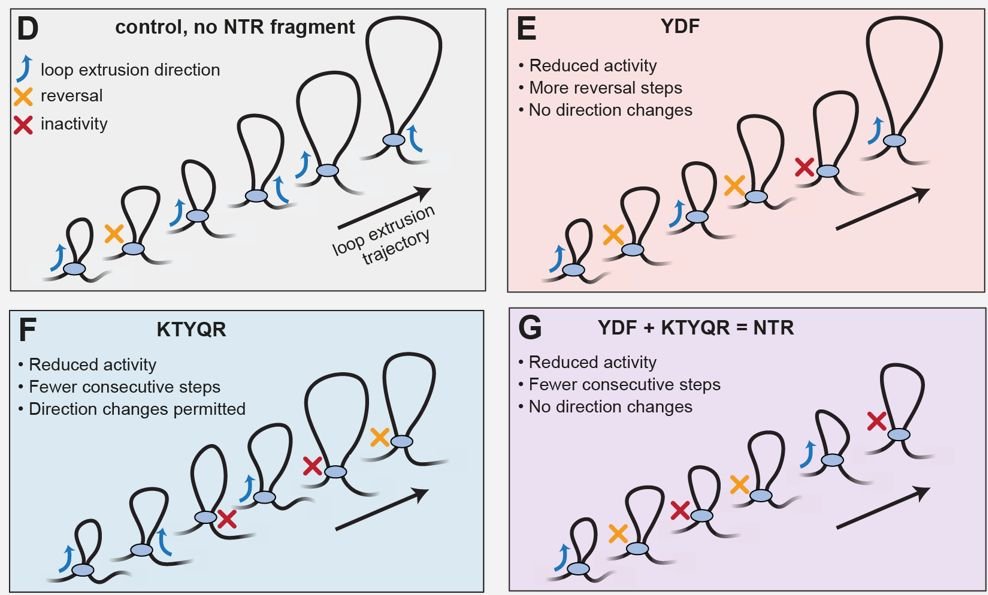
KTYQR is distinct from YxF: it reduces cohesin's LE activity, but doesn't impact its directionality. Together, the two account for most of the impact of the full NTR! We hypothesize that connecting the two on one protein chain and some adjacent sequences modulate further.
1
0
3
A computational AlphaFold screen suggested the KTYQR motif, located N-terminally of the YxF, that appears to also bind STAG1.
1
0
2
Even though YxF has a large impact on cohesin's LE activity, it doesn't fully account for the impact of the full N-terminal region of CTCF. So we searched more 🔎
1
0
1
Curiously, we observed that cohesin also changes direction much less frequently in the presence of YxF! With inspiration from Shaltiel et al, 2022, we hypothesized that this is due to a higher DNA affinity of STAG1-kleisin in the presence of YxF
1
0
1
We started with the known YxF motif. In brief, the YxF motif: - reduces cohesin's ATPase rate - reduces LE initiation - reduces the fraction of complete LE steps (in Magnetic Tweezers) So these few amino acids alone already tinker quite a lot with cohesin!
1
0
2
Observing LE by Magnetic Tweezers provides a much more detailed view. We can see individual steps! Previously, we already observed a plethora of phenomena: cohesin often makes full LE steps, as seen by a height decrease of the magnetic bead. But these are also often reversed
1
0
1
Single-molecule fluorescence visualization lets us assess quantitative metrics such as the loop extrusion (LE) initiation rate, LE rate, directionality changes (see https://t.co/dciPuqjDQ4 ..., etc.
cell.com
All eukaryotic SMC motor proteins extrude DNA asymmetrically but may undergo direction switches, suggesting a common loop extrusion mechanism among all eukaryotic SMC proteins while permitting...
1
0
1
We took the CTCF N-terminal region that is already well-known to interact with cohesin and cut it up into smaller pieces. Then, we exposed cohesin to these various fragments in buffer (no DNA binding domain here!) and observed how cohesin handles that.
1
0
2
CTCF stall DNA 🧬 loop ➰ extrusion by cohesin - but exactly how it pulls this off is a 'mechanistic mystery' (to cite @Anders_S_Hansen , Nucleus, 2020). We just preprinted 📜 a new study @biorxivpreprint that provides some answers to this enigma: https://t.co/CYYDP0NvrE
2
25
82
2024 Fellow @RomanBarth2 led a new study advancing our understanding of how "DNA motors" shape our genome and regulate our genes. He led the work during his pre-Fellowship time at @tudelft For his Fellowship Placement, Roman is at the @UW We're excited to see where he takes it
DNA motors found to switch gears🕹️ Biophysicists found that #DNA motors can switch direction, contrary to what was thought to be possible. The discovery is key to understanding how these SMC motors shape our genome and regulate our genes. 🔗Read more: https://t.co/EdNnQLQPFa
0
2
8
DNA motors found to switch gears🕹️ Biophysicists found that #DNA motors can switch direction, contrary to what was thought to be possible. The discovery is key to understanding how these SMC motors shape our genome and regulate our genes. 🔗Read more: https://t.co/EdNnQLQPFa
0
5
11
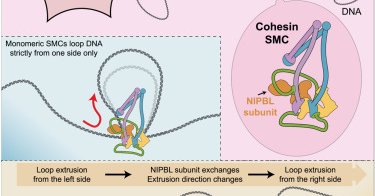
This was another gratifying collaboration between the labs of @Gruber_Lausanne 's and Jan-Michael Peters' @IMPvienna labs. Thank you to all co-authors for your contributions!! We conclude with a fabulous rendering of this mysterious molecular "machine" by the great @Scixel
6
1
2