Rik Lindeboom
@RLindeboom
Followers
651
Following
363
Media
2
Statuses
119
Group leader at Netherlands Cancer Institute / Quantitative Systems Biology
Nederland
Joined September 2011
Exciting news for aspiring PhD students: the NKI PhD Program 2025 will open soon for applications! We participate with multiple top labs in this prestigious program, which offers a unique opportunity for PhD candidates to engage in cutting-edge genome biology and cancer research.
📢 Applications for the NKI PhD program open from Jan 1 - Feb 28. Join our international research community in Amsterdam, working in cancer biology, AI research & life sciences with leading scientists and top facilities #PhD #CancerResearch More info:
0
4
10
Passionate about doing innovative, curiosity-driven research and looking to start your own lab? The NKI has an opening for a Junior Group Leader position! Join us in our collaborative environment in Amsterdam, to use our state-of-the-art infrastructure to make a real impact.
👀 We're looking for a junior group leader! Join our innovative, curiosity-driven institute in a collaborative, international environment. Full freedom to pursue your research, competitive salary & relocation support. 📅 Apply by Dec 1.Info & application:
0
3
9
Happy and proud to receive the ERC Starting grant! We will investigate how genome-wide binding kinetics and energy landscapes of oncogenic transcription factors are regulated!. Interested in joining our team? Apply before the 22th of Sept. #ERCStG.
5
9
60
Excited to share that we received the Technology Impact Award from @CancerResearch to develop methods to study immune - cancer cell interactions for identifying proteins and antigens that modulate cancer cell killing, and targets for immunotherapy!
cancerresearch.org
Dr. Lindeboom's goal is to discover which antigens are the best targets for therapy, potentially leading to safer and more effective cancer treatments.
2
4
36
This was an amazing team effort by the @Teichlab and @drmarkonikolic lab. Massive thanks to my partners in crime @WorlockKaylee and Lisa Dratva, the @ChrisChiuLab that established the model, the involved teams at @sangerinstitute, @ucl, @NKI_nl, @imperialcollege, @hVIVO_UK. (4/4).
2
0
31
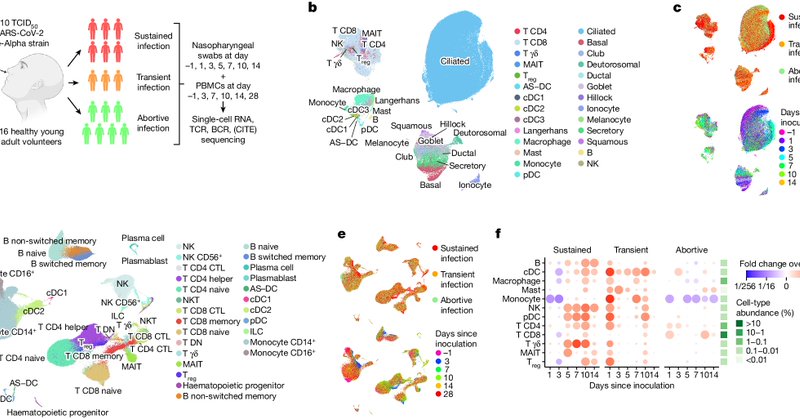
Highlights include the unexpected timing of the interferon response, new innate responses in unexpected cells and in abortive infections, identification and application of new activated T cell populations, and lower HLA-DQA2 expression as predictor for sustained infections. (3/4)
1
2
34
By applying single cell multiomics to a unique SARS-CoV-2 human challenge model, we uncovered the exact timeline of the local and systemic immune responses to viral exposure. This led to both fundamental and therapeutic insights into how our immune system works. (2/4).
1
0
30
Extremely excited to see our paper out in @Nature!. How does the body respond to SARS-CoV-2, and what happens if you don't get infected after exposure?. We unravel the exact timeline of the cellular responses to inoculation with SARS-CoV-2 in humans. (1/4)
nature.com
Nature - A human SARS-CoV-2 challenge study in individuals without previous exposure to the virus or vaccines provides detailed profiles of local and systemic epithelial and immune cell response...
15
116
415
I'm #hiring a #postdoc to join my research group at the NKI in Amsterdam. Are you interested in developing state-of-the-art genomics and proteomics technologies to study TCR : antigen interactions in cancer? Apply before the 8th of May: #AcademicTwitter.
0
4
12
Honored to be among the recipients of the NWO Open Competition M1 grant! We will develop tools to improve our understanding of TCR-antigen interactions, using high-throughput screening & AI. Stay tuned for a postdoc vacancy to join this exciting project!.
nwo.nl
The NWO Domain Board Science has approved nineteen grant applications in the Open Competition Domain Science-M programme. The topics vary from the consequences of hybridization within sea turtles,...
0
4
22
RT @Nature: Nature research paper: Decoding chromatin states by proteomic profiling of nucleosome readers
nature.com
Nature - A multidimensional proteomics analysis of the interactions between around 2,000 nuclear proteins and over 80 modified dinucleosomes representing promoter, enhancer and heterochromatin...
0
35
0
Stoked to see this tour-the-force from the Bartke team out in Nature! We've decoded how DNA & histone modifications regulate protein binding to the genome using quantitative interaction proteomics. A leap forward in understanding chromatin dynamics! Congrats to everyone involved!.
“If you want to know someone, look at their friends…” that’s how we decode histone codes and chromatin states. Finally, the dream has come true… Almost a decade after I started the project in London, it has been published on Nature. Enjoy the paper:
0
1
13
Thrilled to share my journey as a new junior PI at @NKI_nl, featured in @NatureCancer. From overcoming the challenges of setting up a lab to building a team and plotting a scientific course.
💫ONLINE NOW @NatureCancer – the #2023Generation – 12 group leaders share their journey to becoming an independent PI. Very inspiring stories, please do not miss them! .
0
1
26
RT @LaskerFDN: Congratulations to Piet Borst, 2023 #LaskerAward winner! – “for an exceptional 50-year career of scientific discovery, mento….
0
37
0
The NKI opened an exciting new call and PhD program for students interested in Chromosome Biology and Gene Regulation!.
🎓We are looking for outstanding candidates who want to become the new generation of ambitious #PhDstudents in a lively international research community. Check our website to learn about our #PhDprogram in #ChromosomeBiology & #GeneRegulation . ➡️
0
1
7
RT @mnoursad: Do different host transcriptional biomarkers of acute viral infection offer different clinical and research applications? Ins….
medrxiv.org
Evaluation of host-response blood transcriptional signatures of viral infection have so far failed to test whether these biomarkers reflect different biological processes that may be leveraged for...
0
10
0
RT @Schumacher_lab: Excited to share STAPLER, a language model to predict TCR – pMHC reactivity that outperforms prior models. And for ML….
biorxiv.org
The prediction of peptide-MHC (pMHC) recognition by αβ T-cell receptors (TCRs) remains a major biomedical challenge. Here, we develop STAPLER (Shared TCR And Peptide Language bidirectional Encoder...
0
37
0
RT @WorlockKaylee: Our pre-print is out!!. Proud to share our work as part of the world’s first human #COVID-19 challenge study, in which w….
0
19
0
An amazing team effort by the @Teichlab and @drmarkonikolic lab, especially my partners in crime @WorlockKaylee and Lisa Dratva, and @ChrisChiuLab for establishing the model. With teams from @sangerinstitute, @ucl, @imperialcollege, @hVIVO_UK. Thanks to everyone involved! (6/6).
0
0
4