Matthias Vorländer
@MVorlandr
Followers
336
Following
642
Media
21
Statuses
192
Post-doctoral researcher at IMP Vienna
Vienna, Austria
Joined July 2009
New paper alert! Scientists in Clemens Plaschka’s lab at the IMP and Julius Brennecke's lab at @IMBA_Vienna solved a decade-old puzzle, uncovering how the information molecule mRNA travels from the cell’s nucleus to its periphery. More: https://t.co/oKV4XvsYbJ
1
12
52
Thrilled to share that I’ll be joining IMB Mainz in February 2026 to start my own group! We will explore new mechanisms in eukaryotic gene expression, leveraging ‘evolutionary play’ to uncover how regulation, repurposing, and hijacking shape RNA biology. PhD positions available!
1
7
17
In 2019-2021 @BrentNannenga and I put together this methods in cryoem book. It was a lot of work. It sells for 120$ and chapter downloads are 35$. The publisher informed us that they had 78,000 downloads so far. Thats several million dollars profit for them... We got nothing 😳
5
7
64
I'm extremely excited to announce that I've been promoted to Senior Group Leader (with tenure) at the @IMPvienna. A BIG thank you to every lab member for your fantastic work over the years, and to our wonderful colleagues, mentors, and friends for their continued support.
Congratulations to Clemens Plaschka, who was promoted to Senior Group Leader at the IMP. We are thrilled and look forward to Clemens steering his research as part of our community for many more years to come! https://t.co/asQC5kWGaE
13
4
168
We are very excited to share our recent preprint on the nuclear import mechanism of the proteasome, driven by the multivalent adaptor AKIRIN2! https://t.co/dJdiL7RAK8 🧵 (1/7)
biorxiv.org
Nuclear protein homeostasis, including the turnover of transcription factors, critically depends on nuclear proteasomes. After each cell division, proteasomes need to be re-imported into the newly...
1
19
69
The second of our two new preprints addresses the question of how proteasomes can be kept dormant for instance in oocytes. This study was spearheaded by first author Sascha Amann. https://t.co/nBIXIXpTmK (1/6)🧵
2
4
16
🚨Hiring a Research Technician!🚨 Join our lab to study mobile DNA and its impact on biology and genome engineering! 🌟 MSc. in Molecular Biology, Biochemistry or related field required. PhD highly desired. 🌟 70% Research 30% Admin 🌍 @MaxPerutzLabs, @viennabiocenter #Austria
2
62
143
Come and explore the mechanisms of gene expression with us! We are hiring two postdocs in cryo-EM and - with @BalzarottiFran - in super-resolution microscopy. 👉 https://t.co/ipeZ4iQigE 👉 https://t.co/XXlCLZUdBC
imp.ac.at
0
10
23
My favourite discovery ever has just come online. Can I please tell you about some seriously wacky molecular biology? The story starts with a reverse transcriptase that SOMEHOW defends bacteria from viruses. (👇 I recommend sound ON for the video 🎹) 1/
103
1K
5K
Science in Shorts Spotlight: @rooij_laura @PhilDexheimer explain the research on rejuvenating endothelial cells lining blood vessels, which age and cause cardiovascular issues. https://t.co/1JxFoVHjhi
0
3
11
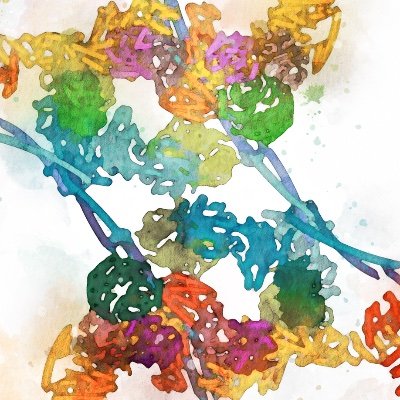
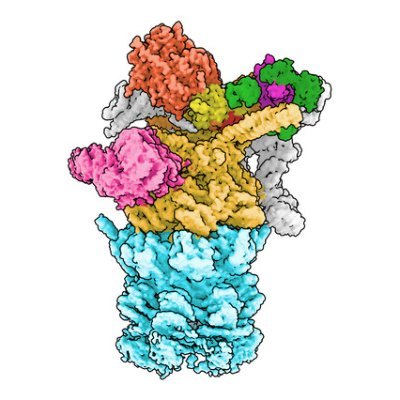
Just released in #EMDB & @PDBeurope and published in @naturemagazine: The composite map of C. elegans Intron Lariat Spliceosome (ILS’) primed for disassembly @ 2.9 Å! The focused maps used to construct this volume are well-linked to this entry.
2
7
35
For the science, check out the paper here https://t.co/gqdkhsjZuS. Many thanks to @PhilDexheimer and the many wonderful colleagues at @plaschka_lab, @LCochella lab and the @IMPvienna who have contributed to this story.
0
2
2
While retaining the molecular detailed captured in our electron microscopy images, Philipp beautifully brought the scene to live by rendering it as if made from wool, capturing the idea of "unravelling" a complex machine from the inside out.
1
0
1
The golden thread symbolizes the catalytic center U6 snRNA, which forms the heart of the spliceosome. The hand symbolizes the RNA helicase DHX15, which initiates disassembly by pulling on this central thread from afar.
1
0
1
Disassembly of the spliceosome by the RNA helicase DHX15 - check out this stunning illustration of our findings published in yesterdays print edition of @nature by the amazing @PhilDexheimer. 🧵
2
7
34
Thank you Sasha and Rui for highlighting our work on the initiation of spliceosome disassembly! https://t.co/WXVhH7gpFw
0
6
20
We previously showed that a coincident FM-FIB-SEM can be turned into a 4D-STEM system for in situ thickness determination ( https://t.co/GKKRmcVJBR). Daan applied this to cryogenic samples, allowing automated, thickness-controlled milling of frozen-hydrated lamella from cells.
1
10
44
Congratulations to winning the 2024 Eppendorf Young Investigator award @plaschka_lab! Clemens is not only a brilliant scientist with a clear vision for his lab, he is also an incredibly supportive mentor and great supervisor. Well deserved!
🎉Congratulations to Clemens Plaschka @plaschka_lab on receiving the 2024 Eppendorf Award! Plaschka is being recognised for his groundbreaking research on the molecular mechanisms of messenger RNA maturation. Full story: https://t.co/ciBwbaPMZi
0
3
27
A big thank you to everyone in this amazing team, the fantastic supervision by @PlaschkaLab and @LCochella, our fantastic colleagues and core facilities at the @IMPvienna and greater @VBC, and @PhDexheimer for some amazing artwork (coming soon! Paper:
1
0
5
Here is a visual summary of these late steps of splicing, starting from the post-catalytic spliceosome and ending with its disassembly (note that how disassembly completes remains unclear)
1
0
1