Lu Lab_Columbia
@LuLaboratory
Followers
755
Following
2K
Media
12
Statuses
352
Chromatin & Cancer Biologists at Columbia university
New York, NY
Joined October 2017
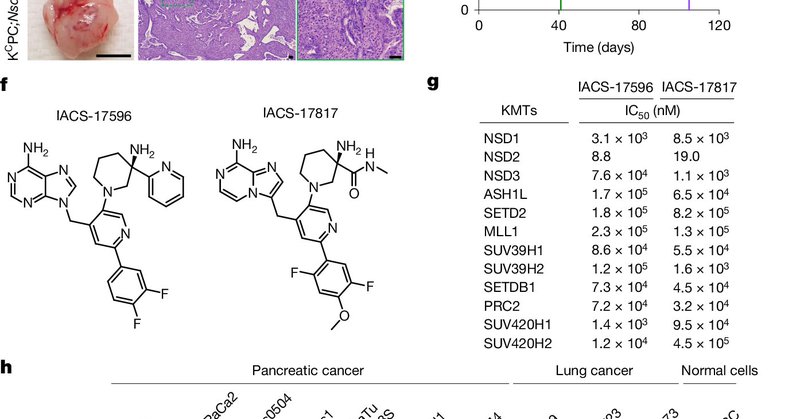
Further, recent work from Gozani, Mazur, Majewski and Jaremko labs demonstrates therapeutic benefit of NSD2 inhibitor in preclinical models of lung and pancreatic cancers, suggesting a broad role of H3K36me2 in cancer chromatin plasticity:
nature.com
Nature - A highly potent and selective small-molecule catalytic inhibitor of the protein lysine methyltransferase NSD2 shows therapeutic efficacy in preclinical models of KRAS-driven pancreatic...
0
0
0
While there is still a lot to learn about the underlying mechanism, these findings suggest the therapeutic potential of combining NSD2 inhibitor with AR inhibitor in treating aggressive prostate tumors. A clinical study is already ongoing:
1
0
0
Here we go! Excited to share our latest collaborative work with @MichaelMShen and @califano_lab. We uncovered that NSD2-catalyzed H3K36me2, a histone mark dearest to our lab, is critical for lineage plasticity and resistance to anti-androgen in prostate cancer.
Nature research paper: NSD2 targeting reverses plasticity and drug resistance in prostate cancer https://t.co/Jbmh0mYAOZ
2
1
5
Want to work with us on DNA methylation and rare genetic disease? Fully funded PhD project with deadline 16th May: https://t.co/ZckwqVoy4M Excited to collaborate with @Hannah_K_Long and @bunina_daria (@EdinUni_IGC @MDC_Berlin). Please share 🙏 #epigenetics
findaphd.com
PhD Project - Fully Funded PhD Studentship in Human Genetics, Genomics and Disease: Dissecting DNMT3B functions in Immunodeficiency-centromeric instability facial anomalies syndrome at University of...
1
5
11
Excited to share this review on chromatin-metabolism crosstalk, lead by our star student Varun Sahu. Here, Varun summarizes the molecular principles governing metabolite-chromatin interaction & highlights emerging technologies that could advance the field. There’re lots to learn!
In this review for our focus issue on patterns in molecular biology, @LuLaboratory explores how metabolites and metabolic enzymes regulate chromatin and histone modification patterns, highlighting the crosstalk between the metabolome and the epigenome. https://t.co/wRBWwxFBif
1
7
41
Check out this important work - congrats to KJ, Victoria and the team!
0
0
2
Please Repost- Excited to be recruiting @CeiFccc @FoxChaseCancer #epigenetic #faculty to continue building our energetic, interactive, supportive #collegial environment Our goal- Basic Discovery & help translate to positively impact patients Apply here- https://t.co/UwItg5JvBN
jobs.sciencecareers.org
The Fox Chase Cancer Center invites applicants for tenure-track faculty positions at the level of assistant professor and associate professor...
0
8
12
Lastly, shoutout to two other papers that characterize the interaction between DNMT3A and H2AK119Ub nucleosome. https://t.co/O7NaCPGmVK by Jikui Song and Greg Wang groups. https://t.co/AoXYF5TUnA by @mdrwilson group.
0
0
1
Congrats again to @KristjanGretars @Xagbomson Susan Gloor, Dan Weinberg, @KJArmache @GCloner @epicypher and everyone involved! Thanks to the reviewers and editor @ScienceAdvances for improving our manuscript.
2
0
2
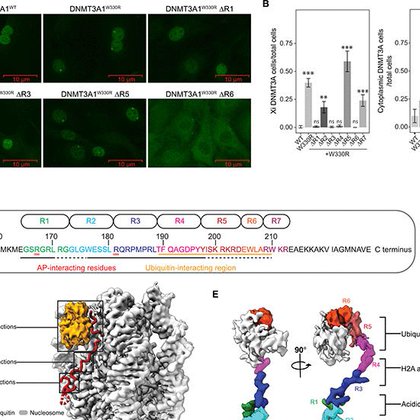
Here we go - our preprint on the interplay between DNMT3A and polycomb H2AK119Ub mark is published. We believe these findings offer a potential mechanism driving DNA hypermethylation in cancer. Pls see tweetorial for more details.
science.org
A structural and genomic analysis of the DNMT3A1 interaction with H2AK119Ub nucleosomes explains its mistargeting in cancer.
Excited to share our new preprint on the interaction between DNMT3A1 and H2AK119Ub. This work is the fruit of a collaboration between @LuLaboratory, @KJArmache Lab and @epicypher and co led with @Xagbomson
5
9
43
What's new and what's next in cancer epigenetics research? Join us for the 2024 VAI Epigenetics Symposium. Our allstar speaker lineup will highlight the latest advances with a focus on mechanisms and therapeutic applications. Registration closes tomorrow! https://t.co/MZsGSckbr0
0
9
21
Please see tweetorial above for more details! As always, we welcome your feedback and suggestions.
0
0
0
It also implies that perturbing the interaction between DNMT3A-H2AK119Ub could be a potential strategy to prevent or reverse DNA hypermethylation in cancer.
1
0
0
Our results suggest that polycomb CGI methylation state reflects the balance between DNMT3A and TET activity. In cancers, either insufficient TET activity, or increased DNMT3A binding, can lead to CGI hypermethylation.
1
0
0
Please see tweetorial below for the latest from the lab led by @KristjanGretars. In collaboration with @GCloner @KJArmache @Xagbomson, this work reveals molecular details of DNMT3A targeting to polycomb CGIs and its implication for cancer-associated DNA hypermethylation.
Excited to share our new preprint on the interaction between DNMT3A1 and H2AK119Ub. This work is the fruit of a collaboration between @LuLaboratory, @KJArmache Lab and @epicypher and co led with @Xagbomson
1
4
27
Free Hybrid World Class Symposium- Featuring Academic and Pharma/Biotech Leaders! Register asap. >400 registered and counting. #Epigenetics #3DGenome #Chromatin
@GAlmouzni @KharasLab @METorresPadilla @ranigeorgelab @LuLaboratory @job_dekker @DaiichiSankyoUS @AstraZeneca @CeiFccc
0
8
15
We thank the editor and reviewers for suggestions of improving our work. And as always, appreciate your feedback!
0
0
0