Morillon’s lab
@LabMorillon
Followers
327
Following
363
Media
46
Statuses
263
lncRNA and dark genome. Created in 2005, now at Institut Curie. tweets from team members. https://t.co/2gYLgGA12a
Paris, France
Joined April 2022
RT @wery_maxime: Thrilled and proud to see the "mRNA Decay: Methods & Protocols" book published. A great and enriching first editorial expe….
link.springer.com
0
14
0
RT @VallotCeline: 📝Epigenomic heterogeneity as a source of tumour evolution. Just out in Nature Reviews Cancer - a piece we wrote with @Mat….
nature.com
Nature Reviews Cancer - Single-cell epigenomic technologies are refining our understanding of cancer evolution. Here Laisné et al. describe how epigenomic heterogeneity generates dynamic...
0
25
0
RT @PPapoutsoglou: New preprint from our lab!! EV-associated lncRNA in cancer! Read below and comment on the content! @LabMorillon @Unit324….
0
2
0
RT @CRougeulle: An important and emotional day to celebrate exciting science in a vibrant atmosphere 🎉. Thanks to the organization team and….
0
3
0
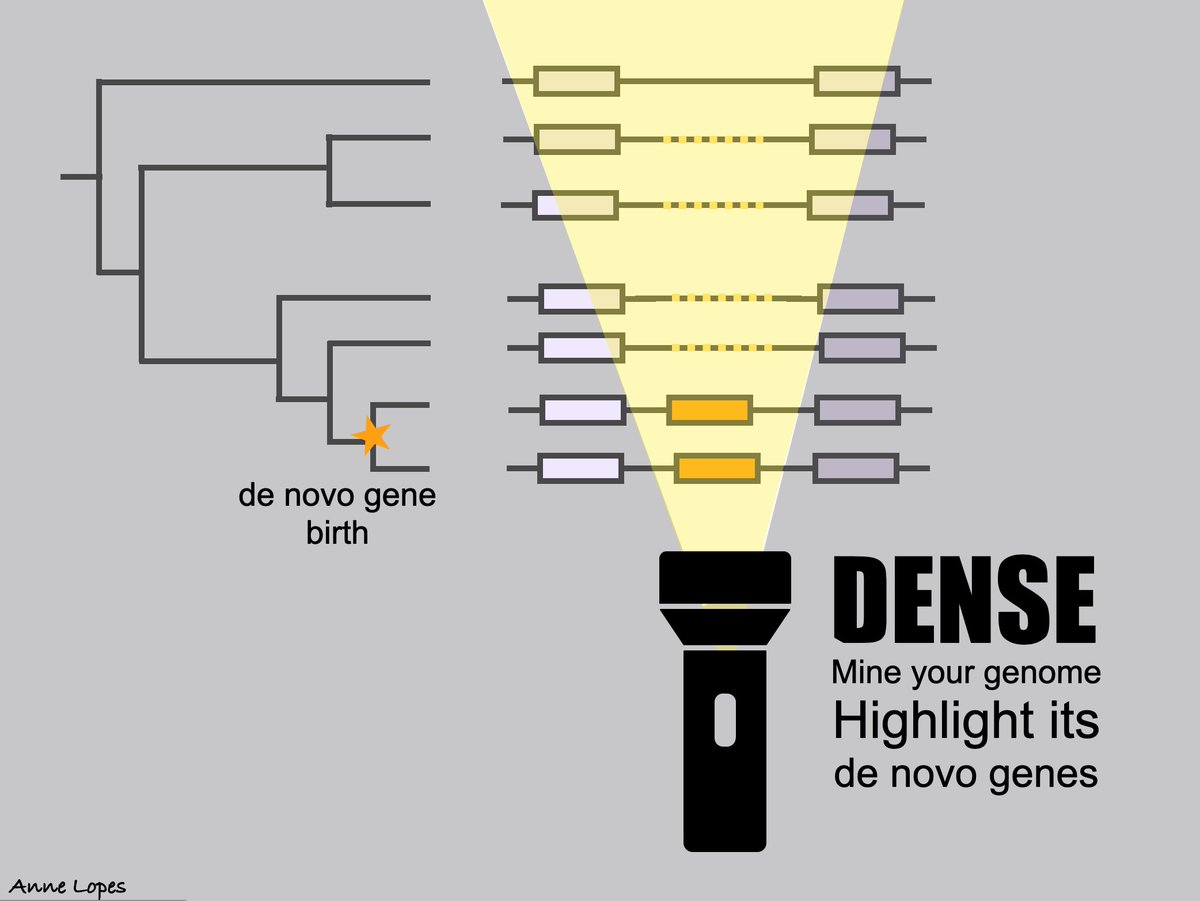
RT @anne__lopes: DENSE is out and ready to mine your favorite genome for its de novo genes! Try it out!
academic.oup.com
Abstract. The discovery of de novo emerged genes, originating from previously noncoding DNA regions, challenges traditional views of species evolution. Ind
0
21
0
RT @RNAJournal: Yeast Xrn1-sensitive unstable lncRNAs contain ribosome-associated small ORFs and are pervasively translated before being de….
0
6
0
RT @vakirlis: I am now looking for a postdoc (or a soon to be one) to apply for a Calmette & Yersin postdoctoral grant to work on this proj….
pasteur.fr
Each year the Institut Pasteur Department of International Affairs allocates funding to promote and aid completion of postdoctoral internships in in one of the 30 Institutes members of the Pasteur...
0
6
0
RT @wery_maxime: Happy to submit the full content of the book "mRNA Decay: Methods & Protocols" that I am editing for the Methods in Molecu….
0
2
0
RT @wery_maxime: Let's meet tonight at poster 522 during poster session 3 of #RNA2024 to discuss about how we should call long noncoding th….
0
4
0
RT @jurivosecchi: Thanks to @institut_curie for the opportunity to present our work in @LabCusanelli at the Non-Coding Genome 2024 Course.….
0
7
0
RT @wery_maxime: @RNAJournal Happy to have our paper published in the June issue, which allows people to see at our work not only at my pos….
0
1
0
The Curie Non-Coding Genome 2024 course is starting today with our fantastic keynote speaker @carninci from Riken institute and human technopole in Milan. Nicely introduced by @MaximeWery @institut_curie
0
3
12
RT @dogranavneet: ~50 years of publication history on #Exosomes or #ExtracellularVesicles #EVs .@IsevComms @IsevOrg #ISEV2024. Data source:….
0
10
0
RT @ScienceCQFD: Et demain 16h ? Reproduire des organes miniatures pour les étudier in vitro , c'est la promesse des organoïdes. sur puce….
0
10
0
RT @DGautheret: You like bad, wicked, aberrant RNAs and want to find them in cancer cell lines: we present our online REINDEER server that….
0
8
0