Hofmann_Lab
@LabHofmann
Followers
164
Following
15
Media
0
Statuses
56
Research group at the University of Cologne
Joined July 2020
Our paper on bacterial clippases is now out in @MolecularCell. These really interesting enzymes can make deconjugation of #ubiquitin irreversible. Thanks to @UniCologne and @C2fBiochemistry for support.
2
21
54
RT @mpimoph: New Parkinson’s disease paper:.Chimeric deubiquitinase engineering reveals structural basis for specific inhibition of USP30 a….
0
3
0
RT @NikolasKlink: Excited to share that our work on Parkinson's target USP30 has been published in Nature Structural & Molecular Biology! B….
0
2
0
Congratulations, great paper! But what makes you think that this is a CARD and not just any death-fold domain? pyrin and DEDs can also activate caspases and invertebrates use all kinds of death-fold domains for that purpose! I don't think this one really should be called CARD.
We previously showed that bacteria encode the pore-forming protein gasdermin and that it defends against phage infection .BUT the mechanism by which the gasdermin system defends bacteria against phage remained unknown.
0
0
1
Great paper, but sorry, this is *not* a CARD domain! It is a 6-helix death fold domain for sure, but there in no evidence provided that it belongs to the CARD subfamily. It might have (convergently?) acquired a similar function as mammalian caspase-associated CARDs. Just saying.
Our paper out @Nature: CARD domains mediate anti-phage defense in bacterial gasdermin systems. CARDs are essential for caspase recruitment in inflammasome activation. We now find them in bacterial immune systems. Congrats @TanaWein! Thanks Kranzusch lab!.
0
1
1
RT @MalteGersch: Very excited to share that our manuscript on the "Discovery and mechanism of K63-linkage-directed deubiquitinase activity….
0
15
0
RT @oleschmoeker: New Lammers Lab paper out now in @NatureComms!🥳.We reveal high functional diversity within bacterial deac(et)ylases by sy….
0
5
0
RT @Jane_Dudley_Lab: Endlessly fascinating and exciting conference with great discussions #EMBOubiquitin24 ! Can’t wait to get back to the….
0
1
0
After a long time in the making, our study on bacterial clippases is finally available as a preprint These are really fascinating enzymes, for which there will be a ton of applications in studying the #ubiquitin system.
2
19
34
RT @MalteGersch: Recent reports implicated USP53 mutations in a hereditary liver disorder in children. We got interested in USP53 and its….
0
1
0
N4BP1 functions as a dimerization-dependent linear #ubiquitin reader which regulates TNF signalling.
0
1
3
Two PhD positions in our research group! If you are interested to study the molecular mechanisms of necroptotic and pyroptotic cell death - and their relationship to plant cell death, do apply!.
Do you love #genetics and have a #MSc in #biochemistry, #biology, or #biophysics? 🧬🌱Join @UniCologne in Germany as #research assistant in the @dfg_public-funded network @sfb_1403 on cell death in immunity, inflammation & disease! Apply by 04/02:
0
6
6
By the way: this project was the brainchild of excellent postdoc Thomas Hermanns, who was also corresponding author! We also acknowlegde support from @C2fBiochemistry and @UniCologne.
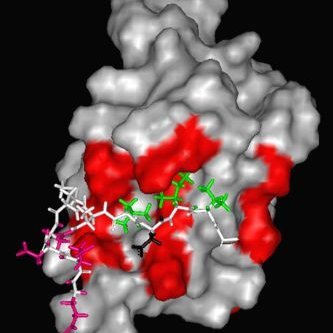
Now published: We analyzed the structure of the Burkholderia effector TssM with and without ubiquitin. TssM is a divergent member of the USP-type deubiquinases and can recognize #ubiquitin despite the lack of the 'fingers' domain.
1
1
5
Now published: We analyzed the structure of the Burkholderia effector TssM with and without ubiquitin. TssM is a divergent member of the USP-type deubiquinases and can recognize #ubiquitin despite the lack of the 'fingers' domain.
1
3
14
Congratulations, Tyler & Jonathan, great work!.
Excited to share the final product of this work, now available at @MolecularCell. Extremely proud of @tylergfranklin for this and many other achievements during his thesis work!.
1
0
2
Out now in NatComms: A Chlamydia-like bacterium with a deubiquitinase repertoire similar to Legionella, but using new DUB classes for the same linkage specificities (e.g. M1, K6). #ubiquitin @UniCologne. @C2fBiochemistry .
1
8
26
Our new manuscript on Burkholderia TssM is on bioRxiv. TssM is an interesting bacterial USP-type dubiquitinase effector, which lacks the so-called 'Fingers' Region used by other USPs for #ubiquitin recognition. Our structures show how TssM does the trick
0
1
2
Nice poster!.
last weeks @BiochemSoc conference on #DUBs and #Ubls in Edinburgh was a blast! lots of new and interesting aspects all around #ubiquitin erasers from molecular basics to applications. thanks to all organizers for putting together this great #biochemevent, see you next time! 🤩
1
0
2
RT @BiochemSoc: DUBS 2023 will bring together leading academic researchers in the DUB/ULP area with their counterparts in biotech and pharm….
0
6
0









