José Jiménez-Gómez
@JimenezGomezLab
Followers
424
Following
1K
Media
31
Statuses
712
Plants research. Head of the Adaptive Genetics and Genomic Lab at CBGP, Madrid. Mastodon: @[email protected]
Madrid, Spain
Joined June 2008
Save the date, 1-3 July 2026, for the XVIII Plant Molecular Biology Meeting #RBMP2026 in Bilbao. Check out the website for updates https://t.co/oq4I1y9X19 Please RT.
rbmp2026.com
RBMP is a key biennial event in the field of plant molecular biology, bringing together researchers, academics, and professionals from across Spain and beyon...
1
25
38
Congratulation to all authors! @Laskotillean @ZeyunXue @Elo1778 @medinajoaquin1 (sorry if I did not find you in X) Institutions @cbgpmadrid
@ijpb_fr
@LIPME_Toulouse
@INRAE_Intl
#GAFL
@CSIC and funding agencies @AgenceRecherche
@AgEInves
1
0
1
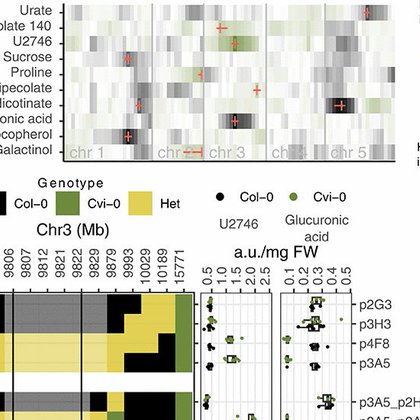
Parallel evolution in this case could be indicative of positive selection to produce the metabolite in this environment. Indeed, plants carrying different alleles of GH38cv present different accumulation of the metabolite, and germinate, and grow better under high salt.
1
0
1
Looking at genome sequences from a global set of Arabidopsis plants (~1600), only two populations have non-functional aleles of GH38cv, and both come from the Cape Verde archipelago, but each population comes from a different island! This is a case of parallel evolution!
1
0
0
We performed metabolite QTL analysis with plants from Cape Verde Islands (Cvi-0) and central Europe (Col-0). The strongest QTL is in an unidentified metabolite. We characterize the metabolite as glucuronyl-mannose, and we map the gene to a glycoside hydrolase family 38 (GH38cv)
1
0
0
We just published our work in #ScienceAdvances "Parallel evolution of salinity tolerance in Arabidopsis thaliana accessions from Cape Verde Islands" https://t.co/0pzTHKGe17 Short summary and credits follow:
2
7
20
Parallel evolution of salinity tolerance in Arabidopsis thaliana accessions from Cape Verde Islands @ScienceAdvances Beautiful work by @Laskotillean and colleagues
science.org
Plants from the Cape Verde Islands evolved parallel mutations to protect from high salinity.
0
10
22
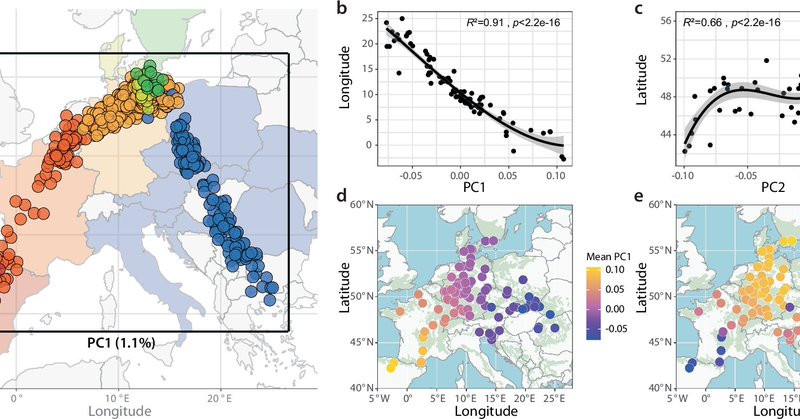
Here it is - our first publication on population genomics of European beech, finally out in Nature Communications: https://t.co/HfDANIeMvd. Big thanks to everybody involved, especially first author Desanka Lazic @VOdorata!
nature.com
Nature Communications - The genomic basis of local adaptation can contribute to predicting species’ future maladaptation. This study shows that local adaptation in European beech is...
1
4
10
For this study, we resequenced beech trees from 100 populations across the distribution range. We find that genetics largely mirror geography.
1
1
1
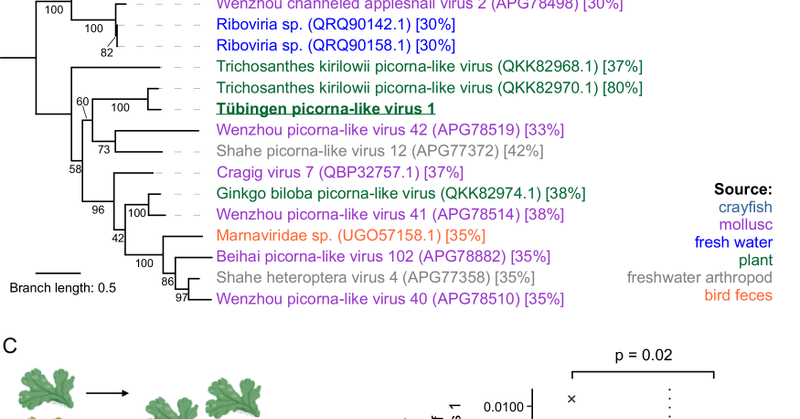
Thrilled to share our study @NatureComm, in collaboration with @DenisKutnjak, on the conservation of the molecular interface between plant and viruses!, we contribute the first comparative study including the non-vascular plant Marchantia polymorpha 🧵 https://t.co/m7KPWizCDf
nature.com
Nature Communications - Assessment of the evolution of plant-virus interactions suggests rapid reshaping of plant viromes after land colonization, along with rerouting of general defense responses...
32
33
138
Very excited to see our paper finally out @NaturePlants ! The manuscript is fully accessible from here: https://t.co/8qHzbjwJfg
https://t.co/xqfxf1EqPN
6
22
56
1/ 🧬 "Can "jumping genes" create novelty? Sure. But how much of this novelty actually serves a purpose? Excited to share our latest preprint, where we report on an adaptive retrotransposon insertion driving rapid responses to herbicides https://t.co/6OGDcxJ1Xn 🧵
7
50
156
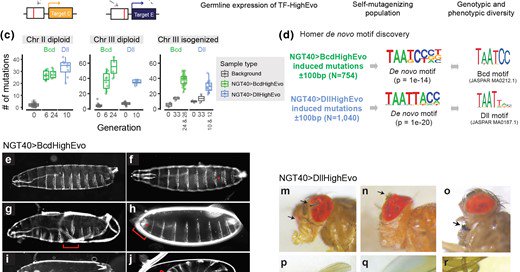
My favorate piece of my postdoc work is published! We fused our favorite TFs with a deaminase domain that induces point mutations in DNA, mutagenizing TF binding sites across the genome. Will be a great reagent for exp. evo., as its name suggests :)
academic.oup.com
Abstract. Understanding the evolutionary potential of mutations in gene regulatory networks is essential to furthering the study of evolution and developme
13
73
360
Great news!! Our paper in the identification of unique #ABA markers for single and #combinedstresses using a #Multiomics approach has been accepted in @JXBot. Great collaboration with the Italian group of Prof. Lucini. @Mirim_science @CEBAS_CSIC
8
6
58
Now published in @ThePlantJournal! Across plant & animal domestications, there was strong selection for visual diversity, creating an incredible palate of colors & patterns. Here, we unravel the genetic/environmental basis of this in common bean! 1/n https://t.co/JXJfYYK11C
3
50
119
Nos ha costado conseguir el proyecto nacional, pero a la tercera va la vencida @JimenezGomezLab! Proyecto del @CienciaGob concedido! @CSIC @CBGP_Madrid
✅Ya puedes consultar la Propuesta de Resolución Provisional de la convocatoria 2023 de Proyectos de Generación de Conocimiento. 🗣️@DianaMorantR: Destinamos 655 M€ a esta convocatoria de investigación, la mayor dotación pública concedida nunca. 📲 https://t.co/SfnKvvFSno
8
2
45
Such a gratifying surprise meeting former MPIPZ colleagues at #XVIIRBMP Castelló 2024. Some are missing in the photo; hopefully next time we can organize better for a selfie! @JimenezGomezLab @fandres1977 @mpipz_cologne
1
3
29
Aristotle was the first to notice honeybees dancing. In 1927 Karl von Frisch decoded the waggle. How it works was "explained" by MV Srinivasan AM FRS in the 1990s. Except @NeuroLuebbert found his papers are junk. A 🧵 about her discovery & our report: https://t.co/K0rpDPnfop 1/
23
333
1K
@TheSainsburyLab @fornaralab @ThorstenLangner @KamounLab Unfortunately last night some people entered the field and destroyed the plants
5
3
20