HittingerLab
@HittingerLab
Followers
2K
Following
505
Media
80
Statuses
610
Biodiversity | Biochemistry | Biotechnology | Functional, Ecological, and Evolutionary Genomics | Moderated by current lab members
Madison, WI
Joined April 2018
It's been awhile since we posted here, but we have an opening for postdoctoral training! Please apply by 15th August 2025 for full consideration. Here is the ad:
0
2
6
For more information and to see our previous winners, see our website @UWEvolution.
evolution.wisc.edu
The J.F. Crow Institute for the Study of Evolution at the University of Wisconsin-Madison is inviting early-career evolutionary biologists from outside UW-Madison to apply to participate in an...
0
0
0
UW-Madison Evolution Seminar Series' Early Career Award is open for applications until November 30th, 2024. This is a great opportunity to showcase your work at our seminar! 🦡⚗️. Postdocs and grad students in evolutionary biology are encouraged to apply:
docs.google.com
Application to the UW-Madison Evolution Seminar Series - Early Career Scientist Awards.
1
1
6
RT @GLBioenergy: 🥐 John Crandall, a postdoc in the @HittingerLab, works with yeasts both in and outside of the @UWEnergy lab as a home brew….
0
2
0
RT @ASBMB: 🍺Q: What makes a yeast variety suitable for lager beer rather than ales? .🍻A: Yeast that ferments best at cool temperatures. Ind….
0
5
0
See the full paper here:. Led by Dr. John Crandall (former @uwgenetics PhD student and current @GLBioenergy postdoc) with Dr. Xiaofan Zhou and @RokasLab.
academic.oup.com
Abstract. Functional innovation at the protein level is a key source of evolutionary novelties. The constraints on functional innovations are likely to be
0
0
2
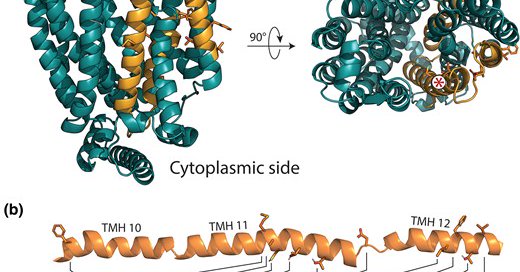
How can a yeast sugar transporter find a new function? .In our new paper out now in @MolBioEvol we look at a recent evolutionary innovation in an alpha-glucoside transporter in S. eubayanus and find that multiple simultaneous mutations are required for this new function!
1
15
60
This work was a great collaborative effort led by @MarieClaireHar2 with @DOpulente @shenxingxing1 @MarizethGroene1 @RokasLab @Lab_LaBella.
0
0
0
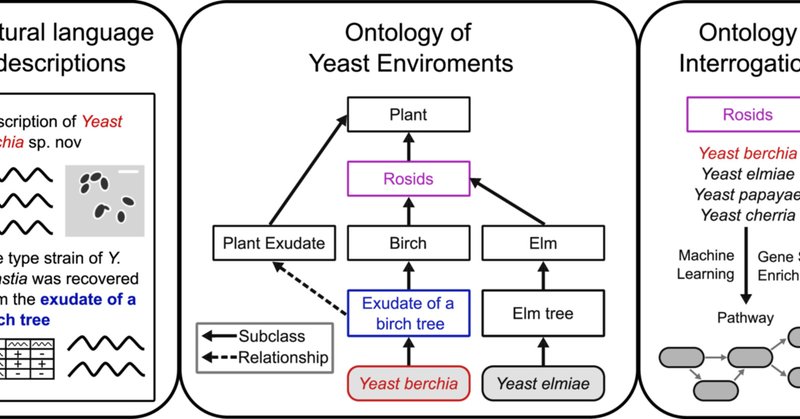
See our new article in which we developed an Ontology of Yeast Environments.🌱This hierarchical framework allows for new systematic analyses and insights into the evolution, ecology, and diversity of yeasts!💡
onlinelibrary.wiley.com
Ontologies are a flexible framework in which we can translate yeast ecological information into a machine-readable format. The creation of the Ontology of Yeast Environments (OYE) allowed us to...
2
14
41
This is based on genome-informed higher ranked classifications led by @MarizethGroene1 as part of the project.
pmc.ncbi.nlm.nih.gov
The subphylum Saccharomycotina is a lineage in the fungal phylum Ascomycota that exhibits levels of genomic diversity similar to those of plants and animals. The Saccharomycotina consist of more than...
0
1
3
Check out @NCBI Insights blog on updating the yeast taxonomy:.
ncbiinsights.ncbi.nlm.nih.gov
As previously announced, NCBI is continually making improvements to our Taxonomy resource in response to new data and changes in biological nomenclature. We recently made classification changes to...
1
9
38
RT @biorxiv_micrbio: Machine learning uncovers gene families impacting oxidative stress resistance across yeasts #….
0
4
0
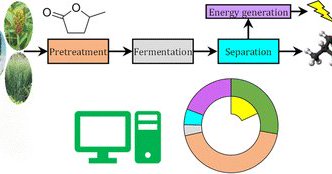
How can yeast fermentation workflows be optimized in biorefineries for isobutanol production?. See our @GLBioenergy collaborative work on using gamma-valerolactone (a renewable green solvent) in biomass pretreatment to achieve high titers of isobutanol.
pubs.rsc.org
The production of isobutanol from lignocellulose has gained attention due to its favorable physical and chemical properties. The use of lignocellulosic biomass as a feedstock to produce isobutanol...
0
3
12
RT @Linder_surprise: Very excited to share this preprint of our work to identify the genetic bases of ROS resistance across the yeast subph….
biorxiv.org
Reactive oxygen species (ROS) are highly reactive molecules encountered by yeasts during routine metabolism and during interactions with other organisms, including host infection. Here, we characte...
0
9
0
How do yeasts resist oxidative stress? We used machine learning to identify gene families that are predictive of ROS resistance across the yeast subphylum. Check out the preprint led by @UWBiochem undergrad Kat Aranguiz and postdoc @Linder_surprise .
biorxiv.org
Reactive oxygen species (ROS) are highly reactive molecules encountered by yeasts during routine metabolism and during interactions with other organisms, including host infection. Here, we characte...
1
13
35