Liping Zhao
@Gutzhao
Followers
2K
Following
312
Media
52
Statuses
642
A microbial ecologist with over thirty years of experience on gut ecology. Nutrition, Microbiome and Health; Reference-free and Guild-based Microbiome Analysis.
Joined July 2015
Your gut microbiome is a seesaw for health! 🦠 Fiber-rich diets tip it toward Foundation Guild dominance, promoting weight control and metabolic health. Junk food fuels Pathobionts, driving obesity. #TCGScience #GutHealth
0
0
3
The Two Competing Guilds (TCG) Model of Core Microbiome
🎙️We interviewed Prof. Liping Zhao (@Gutzhao), Executive Editor of #Microbiome Research Reports and Fellow of the American Academy of Microbiology. 🔗 Watch the full interview: https://t.co/KmjvQbDsYK
#GutHealth #Obesity #MRR #HostMicrobe
0
0
10
🎙️We interviewed Prof. Liping Zhao (@Gutzhao), Executive Editor of #Microbiome Research Reports and Fellow of the American Academy of Microbiology. 🔗 Watch the full interview: https://t.co/KmjvQbDsYK
#GutHealth #Obesity #MRR #HostMicrobe
0
1
7
Taxa should no longer be used as entry point analytical units for microbiome datasets. Genomes should be.
0
0
8
🎉Huge congratulations to our Editor-in-Chief, Dr. Jizhong Zhou, on being elected to the National Academy of Sciences! This prestigious honor recognizes his outstanding contributions to Microbial Ecology. We are proud and thrilled! @theNASciences
https://t.co/gznLIgxDzY
1
7
13
Researchers, including CIFAR's @Gutzhao, have developed a new method to identify crucial gut microbes essential for health. 🦠 This discovery could lead to precision nutrition and personalized therapies for managing diabetes, IBD and more. https://t.co/4yehz8aoIF
medicalxpress.com
Researchers at Rutgers University-New Brunswick, along with international collaborators, have introduced a novel method for identifying the crucial set of gut microbes commonly found in humans and...
0
2
6
Immensely grateful to all experts who contributed this piece with their expertise and efforts! @nsegata @cibiocm @GiovanniCammar9 @h_sokol @fasnicar @Siew_C_Ng @Gutzhao @PaedsRH @RaesLab @HoldGeorgina @ChristianHvas @KorenLab @TheTunLab @MireiaVallesc @mcarmen_collado @epasolli
1
1
11
Meet Evo, the DNA-trained AI that creates genomes from scratch | Science | AAAS
science.org
ChatGPT-like model learns on its own to devise new proteins and genetic sequences
0
0
4
The third pillar of guild-based microbiome data analysis is interaction focused perspective. Co-abundance network analysis of high quality MAGs is used to identify potential guilds
The second pillar of guild-based microbiome data analysis is database independence. Each high quality MAG is given a Universal Unique Identifier (UUID) and tracked in samples across different studies for data integration and mining.
0
0
5
The second pillar of guild-based microbiome data analysis is database independence. Each high quality MAG is given a Universal Unique Identifier (UUID) and tracked in samples across different studies for data integration and mining.
Guild-based analysis of microbiome data has three pillars, the first is genome-specific analysis with high resolution and granularity than any taxa. High quality metagenome-assembled genomes (MAGs) are the basic features.
0
0
2
Guild-based analysis of microbiome data has three pillars, the first is genome-specific analysis with high resolution and granularity than any taxa. High quality metagenome-assembled genomes (MAGs) are the basic features.
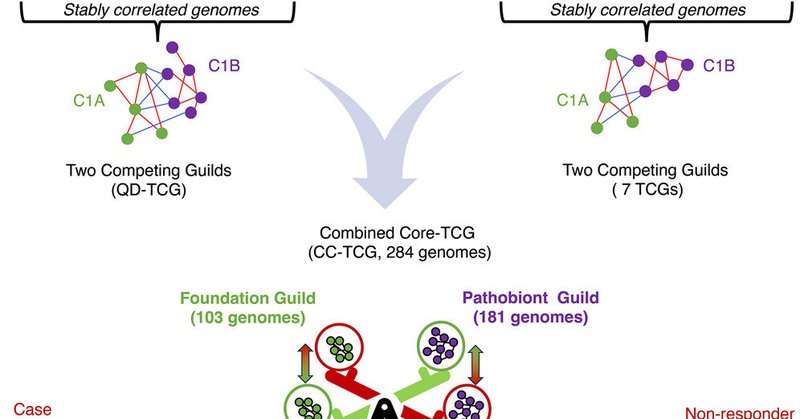
What kind of analytical strategy would allow you to integrate 38 different microbiome studies and identify a set of 284 genomes of core bacteria into Two Competing Guilds as an indicator of Health: the guild-based analysis!
0
0
3
What kind of analytical strategy would allow you to integrate 38 different microbiome studies and identify a set of 284 genomes of core bacteria into Two Competing Guilds as an indicator of Health: the guild-based analysis!
cell.com
This study identifies gut bacteria that maintain stable, enduring relationships under various perturbations, such as dietary changes and disease states. These bacteria form a core microbiome struct...
0
0
6
🚨New Paper Alert🚨 Our new #microbiome research, co-led with @LiatShenhav, published today in @CellCellPress! We show that #breastfeeding "paces" gut & nasal microbiome development to prevent #asthma in the @CHILDSTUDY. #Tweetorial 🧵 https://t.co/eeHSLIdw9Q
9
92
323
This concept of stability of relationships as a marker of functional significance is central to understanding the microbiome as a complex adaptive system, where the resilience of microbial networks is directly linked to host health.
cell.com
This study identifies gut bacteria that maintain stable, enduring relationships under various perturbations, such as dietary changes and disease states. These bacteria form a core microbiome struct...
Are we ready to define a core microbiome signature as an indicator of health? .@Gutzhao et al. suggest a stable interaction-focused approach to enhance our understanding of the fundamental structure of the gut microbiome: https://t.co/11mTTpUfn1
0
0
1
In essence, stability within guilds like FG and PG reflects an evolutionary adaptation, enabling the microbiome to sustain functions under environmental pressures that a less connected population could not.
cell.com
This study identifies gut bacteria that maintain stable, enduring relationships under various perturbations, such as dietary changes and disease states. These bacteria form a core microbiome struct...
Are we ready to define a core microbiome signature as an indicator of health? .@Gutzhao et al. suggest a stable interaction-focused approach to enhance our understanding of the fundamental structure of the gut microbiome: https://t.co/11mTTpUfn1
0
0
1
Restoring and maintaining the TCG structure in the gut microbiome is therefore essential for it to function as a dynamic regulatory interface between the host and the environment.
cell.com
This study identifies gut bacteria that maintain stable, enduring relationships under various perturbations, such as dietary changes and disease states. These bacteria form a core microbiome struct...
Are we ready to define a core microbiome signature as an indicator of health? .@Gutzhao et al. suggest a stable interaction-focused approach to enhance our understanding of the fundamental structure of the gut microbiome: https://t.co/11mTTpUfn1
0
0
1
It illustrates how dietary choices shift the balance toward either health or dysfunction, suggesting that restoring FG dominance through fiber-rich diets could recalibrate this balance, realigning it with the microbiome’s evolutionary purpose.
cell.com
This study identifies gut bacteria that maintain stable, enduring relationships under various perturbations, such as dietary changes and disease states. These bacteria form a core microbiome struct...
Are we ready to define a core microbiome signature as an indicator of health? .@Gutzhao et al. suggest a stable interaction-focused approach to enhance our understanding of the fundamental structure of the gut microbiome: https://t.co/11mTTpUfn1
0
0
2
Thus, the stable competition between FG and PG is fundamental to the microbiome’s role as an adaptive nutrient sensor.
cell.com
This study identifies gut bacteria that maintain stable, enduring relationships under various perturbations, such as dietary changes and disease states. These bacteria form a core microbiome struct...
Are we ready to define a core microbiome signature as an indicator of health? .@Gutzhao et al. suggest a stable interaction-focused approach to enhance our understanding of the fundamental structure of the gut microbiome: https://t.co/11mTTpUfn1
0
0
1
This imbalance exemplifies how the FG-PG competition, although adaptive, becomes maladaptive in today’s dietary landscape, where one guild is consistently favored.
cell.com
This study identifies gut bacteria that maintain stable, enduring relationships under various perturbations, such as dietary changes and disease states. These bacteria form a core microbiome struct...
Are we ready to define a core microbiome signature as an indicator of health? .@Gutzhao et al. suggest a stable interaction-focused approach to enhance our understanding of the fundamental structure of the gut microbiome: https://t.co/11mTTpUfn1
0
0
1