Geoff Faulkner
@Faulkner_Lab
Followers
3K
Following
3K
Media
176
Statuses
2K
Professor, University of Queensland. Retrotransposons and genomics. Views my own. Scholar publications: https://t.co/xQu0hWudj1 @faulknerlab.bsky.social
Brisbane, Queensland
Joined July 2017
After years of telling visitors to Brisbane what an awesome place it is for fun* adventures, I've decided to make a list in the order I remember them of my favourite places to go for ease of reference. Here goes ... (*definition of fun may vary)
6
2
34
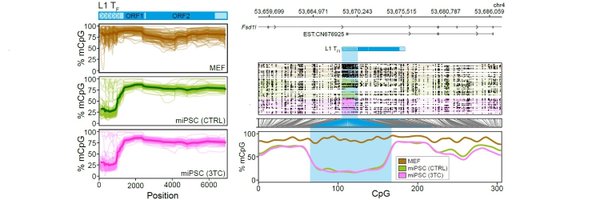
Reposting comment on preprint of this @Dev_Cell paper. Stats done on technical replicates of L1 KD to make small absolute changes seem significant. Biological replicates were available but not used for stats, why? Because p>0.05? I don't understand this. https://t.co/DMsoTLzr3S
I doubt the stated phenotype here is due to changes in L1 mRNA. The ASO and CRISPRi KD approaches (Figs 1, S1, S2) reduce L1 expression by <10% and these small changes are noted as efficient and significant KD. Stats were done on tech reps to give p<0.05 https://t.co/npX6YyPLXq
1
1
17
1/ Our lab’s first preprint found a wonderful journal home! 🎉 Our paper explores the role of testis-specific transcription factors in Drosophila and reveals broad functional connections between tissue-specific paralog proteins. 📝:
5
12
31
Thoughtful and rigorous work from @RippeiH and @PRAndersen teams looking at PAF1C and transcriptional termination. Interesting for me to look at "the other end" of transcription for a change. I wonder what brings tTAF and tPAF together? https://t.co/sryrqH1eWo
1
0
8
🚀 Join the CRG as a Junior Group Leader in Quantitative Cell Biology! 🧬 We’re seeking passionate leaders to tackle fundamental biological questions using cutting-edge multidisciplinary approaches. Enjoy generous core funding, state-of-the-art facilities, and a collaborative
1
27
23
The 5-year postdoc position in my lab is now "live" for applications! Deadline to apply is November 3rd: https://t.co/Mv9zu9t6mT Please RT
imperial.ac.uk
Please note that job descriptions are not exhaustive, and you may be asked to take on additional duties that align with the key responsibilities ment...
Our collaborative NIH-R01 to study the role of TEs in ALS was awarded today, in collaboration with Davide Trotti and Piera Pasinelli (ALS Center at Thomas Jefferson University). Our lab in London will soon advertise a 5-year postdoc position. Please reach out if interested!
0
36
43
I doubt the stated phenotype here is due to changes in L1 mRNA. The ASO and CRISPRi KD approaches (Figs 1, S1, S2) reduce L1 expression by <10% and these small changes are noted as efficient and significant KD. Stats were done on tech reps to give p<0.05 https://t.co/npX6YyPLXq
biorxiv.org
The family of LINE1 transposable elements underwent a massive expansion in mammalian genomes. While traditionally viewed as a mutagenic selfish element, recent studies point to roles for LINE1 in...
0
1
19
Looking for a PhD? Interested in epigenetics, imprinting and transposons? Come join the @the_FrostLab and @rjoakey2012 labs for our project in the 2025 @KCL_MRCDTP scheme, applications are now open! Link to project: https://t.co/vyIyJa6KVB
@KingsCollegeLon @theMRC
#MRCDTP25
1
58
143
I am looking for a motivated PhD student who is interested in understanding the fundamentals of gene regulation. They will be working directly with me on a DECRA funded project (scholarship guaranteed for domestic students). DM me if you know of anyone interested, including you
55
161
392
interested in 3D genome organization during early development? want to understand how it is shaped by RNA? you like data analysis and developing new computational tools? 🧬💻🧐 With @golobor we are recruiting a PhD student/intern to work with us at @IMBA_Vienna
My lab is looking for PhD students with a background in physics and/or biochemistry/quantitative biology. If interested, please apply to the VBC graduate school. Deadline: Oct 15th.
1
14
25
Congrats @JuanManuelBotto for passing your final PhD milestone and welcome to the lab @barunlz !
2
0
15
Delighted that this is finally out! Thanks to @Faulkner_Lab @todd_macfarlan and the other reviewer for good and constructive comments that arrived in my inbox the day after my second child was born... paper here https://t.co/YfN3uIo7XI. Short reflection on new stuff...
nature.com
Nature Communications - Here, the authors investigate the relationship between HUSH-MORC2 and DNA methylation in embryo-derived human neural progenitor cells, enhancing the understanding of the...
Happy and proud to share my lab’s study on how DNA methylation and the HUSH-MORC2 corepressor work together, out last week on biorxiv: https://t.co/lbjxYH1wpS A fantastic collaboration with our colleagues at @JakobssonLab @Lund_Stem...1/7
2
6
31
Calling all emerging scientists - it’s time to send us an application! 😉😎 Lead your own #MaxPlanck Research Group w/ €2.7M in funding over 6 yrs, full independence & resources to hire staff. State-of-the-art infrastructure awaits you!🌟 Apply today➡️ https://t.co/cHxPLyRj9x
mpg.de
Apply to become a Max Planck Research Group Leader. Max Planck Research Groups - Announcement 2024.
9
560
1K
Preprint on "BWT construction and search at the terabase scale". We can compress 100 human genomes to 11GB in 21 hours, find SMEMs with it, do affine-gap alignment and retrieve similar local haplotypes. 7.3Tb commonly sequenced bacterial genomes ⇒ 30GB https://t.co/DiRwZNHVVa
9
222
726
Proud to have contributed to this fantastic study led by @ch_douse . HUSH for the world! https://t.co/dEK1Qoiob7
nature.com
Nature Communications - Here, the authors investigate the relationship between HUSH-MORC2 and DNA methylation in embryo-derived human neural progenitor cells, enhancing the understanding of the...
0
6
23
1/11 Welcome to our August ECR spotlight! This month we get to know Charles Bell @charles_bell92 who works in the Faulkner lab @Faulkner_Lab at the Mater Research Institute @MaterResearch and University of Queensland.
2
2
5
Just under a week to apply to join our team working on the spatial transcriptomics (#MERSCOPE, #VisiumHD, #Xenium, and more..) of poor prognosis cancers. https://t.co/3pWRzpXR9q
0
8
6
Please RT! 🧬🧬 I am looking for a #postdoc fellow to lead a new project in 3D chromatin organisation and transcriptional regulation in cancer (experimental and/or computational) at @SAiGENCI in Adelaide. Apply below: https://t.co/Uxeq1G9iI6
0
88
101
In sum, my alternative hypothesis was not proven and the RNLTR12-int siRNA doesn't seem to impact Mbp. We discussed all of this with the authors as we went along but I also thought there was a good chance someone else might see the same thing - if so there's an answer here. 5/5
0
0
1
Juan made a HEK293T line (where RNLTR12-int should be absent) stably expressing rat Mbp and introduced the RNLTR12-int siRNA, then used the published Mbp qPCR primers to evaluate the siRNA's effect on Mbp compared to a non-targeting siRNA. No significant effect was observed. 4/n
1
0
1
Thinking about the mechanistic model, I wondered if instead the RNLTR12-int siRNA directly targeted Mbp. Aligning the two sequences I noticed the siRNA seed region matched Mbp pretty well. @JuanManuelBotto volunteered to test this possible off-target match experimentally. 3/n
1
0
2