Ellie Rand
@EllieRand3
Followers
144
Following
132
Media
6
Statuses
39
Harvard Systems Biology PhD in the @baym lab. Petitioning for a bacteria emoji
Joined December 2019
Get ready to dance, our paper – Phage DisCo: targeted discovery of bacteriophages by co-culture – has been pre-printed! 🪩 This has been a really fun project to work on with @implosian, @baym, @nquinoneso, Kesther and Carmen! https://t.co/k04NdEUvss 1/
biorxiv.org
Phages interact with many components of bacterial physiology from the surface to the cytoplasm. Although there are methods to determine the receptors and intracellular systems a specified phage...
3
31
74
The latest work from my lab, Phage Disco, a method @EllieRand3 developed for targeted discovery of bacteriophages based on the bacterial receptor, defense system, or other component they interact with, is now live in mSystems
2
30
146
.@EllieRand3 (@harvardmed) etal developed a new targeted #Phage Discovery method based on multiple fluorescent markers, allowing for co-culture w/ multiple hosts & visualization of phages — no more selecting only for dominant phages in samples! https://t.co/C58ZOfsQxd
1
9
14
We've long been able to find new phages and figure out what they interact with. But what about the other way: if you have a gene, like an antibiotic resistance efflux pump, or a phage defense system, and you want to find phages that do or do not interact with it? Now you can!
@implosian @baym @nquinoneso What if we dug into a sample a little more “wild” and a little less characterized? We combined our strains with a wastewater sample (thank you Boston for your ~contributions~), and we found fluorescent hits! Each was isolated and characteristics confirmed on monoculture lawns. 5/
1
13
36
@implosian @baym @nquinoneso Thank you so much to everyone who has worked on this project with us! It’s been so much fun to hear about the phages people are interested in isolating. We are so excited about the upcoming collaborations to put this method to good use! 9/9
0
1
5
@implosian @baym @nquinoneso Phage DisCo is a reliable way to target phage discovery. It can be modified to your chosen phage characteristics of interest by selecting which strains to screen on. The only requirements are genetic tractability of the bacterial host and an environmental sample to screen! 8/
1
1
5
@implosian @baym @nquinoneso Yet again, the DisCo ball (fluorescence) led us quickly to phages of interest. The hits were isolated and confirmed by plaque assays on monoculture lawns of each strain. 7/
1
1
5
@implosian @baym @nquinoneso What about other bacterial factors that phages interact with during infection? We co-cultured a WT E. coli with an isogenic strain that had GmrSD, a modification-dependent restriction enzyme, introduced. If GmrSD provides defense against a phage, we expect that strain to grow. 6/
1
1
3
@implosian @baym @nquinoneso What if we dug into a sample a little more “wild” and a little less characterized? We combined our strains with a wastewater sample (thank you Boston for your ~contributions~), and we found fluorescent hits! Each was isolated and characteristics confirmed on monoculture lawns. 5/
2
2
6
@implosian @baym @nquinoneso Phage DisCo can be further multiplexed with three bacterial strains in co-culture, and, when tested with characterized phages, the fluorescent signal shined! Maybe you can see where the disco ball comparison comes in here. 🪩 4/
1
1
5
@implosian @baym @nquinoneso What if, instead of sampling hundreds of phages, we could just make the right ones glow? Enter Phage Discovery by Co-culture (Phage DisCo) which combines two strains: a WT and a modified strain, each fluorescently tagged. Now, fluorescent signal will mark plaques of interest. 3/
1
2
5
@implosian @baym @nquinoneso While it is widely recognized that phage diversity is a valuable resource, techniques to sample phages have not changed in almost 100 years. These traditional methods tend to oversample abundant phages and miss rare phages where most of the diversity is harbored. 2/
1
1
5
Very excited to add this financial support to the educational, research, and emotional support that my mentors, lab mates, friends, and family have provided throughout grad school <3
Huge congratulations to @EllieRand3 for receiving an F31 from NIAID to support her and her thesis project “Receptor-Guided Discovery of Environmental Phages” with me and @PaulTurnerLab!
3
1
20
Living the [phage biologist's] dream today, touring the legendary Deer Island wastewater treatment plant in Boston🥚🥚🥚
1
6
61
Having a great day visiting Boston and rubbing shoulders with @Baym in his lab (where my talented daughter is a PhD candidate 🙂❤️)
0
3
31
Check out this awesome work by @nquinoneso and @implosian on discovering and exploring the world of plasmid dependent phages! It was so fun to watch this project come together, and I learned so much from you guys along the way. Congratulations!!
Exciting news! The pre-print for our paper, “Diverse and abundant viruses exploit conjugative plasmids” is finally out! Join me on this thread to hear more about this mysterious group of phages with rather unique lifestyle preferences. 1/
0
1
5
Today in @Nature we share our work describing • how to achieve resistance to natural viruses, • a technology that prevents the escape of engineered genetic information, • how viruses overcome genetic-code-based resistance. https://t.co/fZlLxWPBky
23
202
671
Cookie decorating competition at the @baym lab retreat using the @ZymoResearch cookie cutter! Thank you for the decoration inspo! Which is your winning cookie?
2
8
33
If you are applying to the @HMS_SysBio SSQBio PhD program this cycle, consider applying to our application assistance program! Participants will be matched with current students who will help with application materials and answer any questions. https://t.co/wXU8P8XpQ2
docs.google.com
The Systems, Synthetic and Quantitative Biology (SSQBio) Application Assistance Program (AAP) is a volunteer-student run program meant to assist eligible applicants to the PhD program for the...
0
2
0
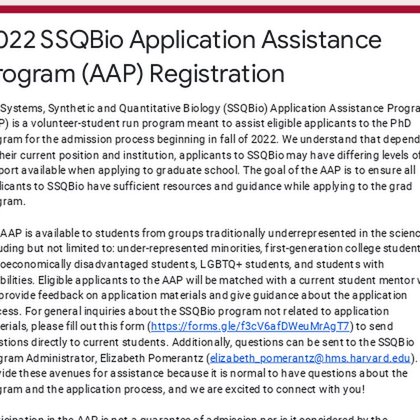
It's time for the 2022 application cycle! We are reopening our @HMS_SysBio SSQBio application assistance program for the third year at the following link. Can't wait to meet the new applicants! https://t.co/GlAjPT5yuA
docs.google.com
The Systems, Synthetic and Quantitative Biology (SSQBio) Application Assistance Program (AAP) is a volunteer-student run program meant to assist eligible applicants to the PhD program for the...
Students in SSQBio @ Harvard want to make sure everyone has access to resources when applying to graduate school, so we created an application assistance program (AAP) and an email helpline to assist current applicants. Links below (1/3)
0
5
7