Beckmann Lab
@BeckmannLab
Followers
2K
Following
793
Media
65
Statuses
171
No longer updating this account. Find us @beckmannlab.🦋.social
München, Bayern
Joined April 2018
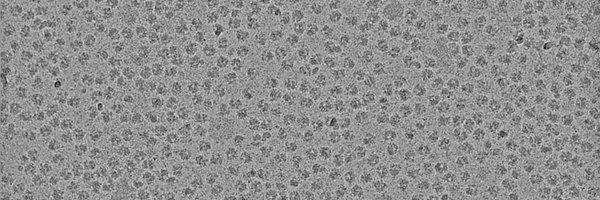
Read Rosales-Hernandez et al. https://t.co/hGCYjaO1nu and Zerbes/Colina-Tenorio et al. https://t.co/3tl0q05sy0 to find out about delivery of the Rieske protein after Bcs1 transport to Mar26 and MICOS for bc1 complex assembly.
embopress.org
imageimageBcs1 is an unusual, heptameric AAA-ATPase that translocates its cargo, Rip1, across the inner mitochondrial membrane in a fully folded state. Cryo-EM and functional data provide novel...
0
2
4
Our results finally explain the essential role of snR30 snoRNA! Check out the details in our preprint to find out more 👉👉👉 https://t.co/UJ8ib8Miw0
#ribosome #cryoEM #RNA
biorxiv.org
During synthesis, modification and maturation of the ribosomal RNA, correct subdomain folding without additional guidance poses a major challenge. An essential H/ACA snoRNP, snR30, was observed to be...
0
7
40
First structure of a H/ACA snoRNP acting in ribosome synthesis. In a great collaboration with the Hurt lab, we provide a detailed structural and biochemical view of the snR30 snoRNP guiding local 18S rRNA subdomain folding. 👇👇👇
2
24
176
Authors including @jingdongcheng report cryo-EM structures revealing how antibiotic tigecycline can also target human mitoribosome. https://t.co/HFoYItNpQl
0
10
23
For a more detailed look, check out the paper now👉
nature.com
Nature Communications - Otu2-driven deubiquitylation of ribosomal protein eS7 impacts translational efficiency. Here, the authors provide the molecular basis for recognition of monoubiquitinated...
0
0
7
Otu2 can bind to 40S during cytoplasmic ribosome biogenesis and all stages between ribosome recycling and start-codon recognition, but becomes a highly specific enzyme for translational reset during recycling/(re)initiation by removing mono-ubiquitin present on eS7.
1
0
3
We also solved the crystal structure of the OTU domain of Otu2 and showed how it recognizes and deubiquitinates eS7 with Lys83-linked monoubiquitin. This binding mode explains the discrimination against 80S ribosomes and the specificity for mono-ubiquitinated eS7 on 40S.
1
0
1
We used cryo-EM to visualize native and reconstituted Otu2-bound ribosomal complexes at different stages of translation. We found that Otu2 binds to 40S subunits mainly between ribosome recycling and initiation stages.
1
0
2
Otu2 is a deubiquitinating enzyme that removes ubiquitin from eS7, a ribosomal protein of the 40S subunit. Ubiquitination of eS7 by Not4 E3 ligase occurs during active translation and affects translation efficiency. However, the role and mechanism of Otu2 are poorly understood.
1
0
2
🎉We are thrilled to share our exciting new collaboration with @inada_lab and @jingdongcheng on how Otu2 recognizes and deubiquitinates 40S ribosomes after translation. Here is a thread with the most interesting findings:👇 #ribosome #ubiquitin #cryoEM
1
17
57
Turns out MutS2 just doesn’t cut it ✂️ despite it having an SMR domain! Our latest collaboration with the @BeckmannLab characterizing the role of MutS2 in splitting stalled ribosomes in B. subtilis. https://t.co/OfAJSzBeOR
biorxiv.org
Stalled ribosomes are rescued by pathways that recycle the ribosome and target the nascent polypeptide for degradation. In E. coli , these pathways are triggered by ribosome collisions through...
0
14
61
Let's rotate! Thanks to the great Hurt lab and @BeckmannLab, our story on the 60S ribosome biogenesis is finally out in EMBO reports. We find the 5S RNP in a new rotate conformation when stably incorporate into the nucleolar pre-60S. #cryoEM #ribosome
https://t.co/w6sIVUGKvw
1
12
41
Its online:🥳🎊🎉 Check out the @MathWorksAI user story🍻 https://t.co/Ikr9VP8TfG about our (@BeckmannLab, @hopfnerlab) DL based Cryo-EM image analysis tool ( https://t.co/wgclFJYrhI). Thank you @MathWorks for the great support!👍👍
nature.com
Scientific Reports - Ice thickness monitoring for cryo-EM grids by interferometry imaging
0
3
9
The Ski2-like helicase 1 subunit applies a pulling force on the mRNA which causes RQT to swivel between two conformations. This leads to the destabilization of the 40S subunit, resulting in subunit dissociation of the leading ribosome.
2
0
3
The ribosome-associated quality control pathway is activated when ribosomes collide during persistent stalling. The RQT complex splits the stalled ribosome, thus clearing harmful traffic jams in the cell.
1
0
0
Keeping up the momentum, we're excited to share the final version of our story about RQT-driven ribosomal splitting 🎉 🎉 Shout out to @KaBe__01, @ThomasB08006066, @inada_lab and everyone involved! Read on to learn more👇🏻👇🏻👇🏻 #cryoEM #ribosome #RQT
Pull the ripcord to clear the traffic jam: RQT splits collided #ribosomes by engaging the emerging mRNA. Check out our new story at #bioRxiv: https://t.co/JhCklVwKvW
2
17
64