Amber Tang
@AmberZqt
Followers
134
Following
21
Media
1
Statuses
16
Research Scientist @instadeepai /Deep Learning for Bio/ genomic LLMs/
Joined August 2015
Our work on "Evaluating the representational power of pre-trained DNA language models for regulatory genomics" led by @AmberZqt with help from @NiraliSomia & @stevenyuyy is finally published in Genome Biology! Check it out! https://t.co/AFBC9Qu4x3
genomebiology.biomedcentral.com
Background The emergence of genomic language models (gLMs) offers an unsupervised approach to learning a wide diversity of cis-regulatory patterns in the non-coding genome without requiring labels of...
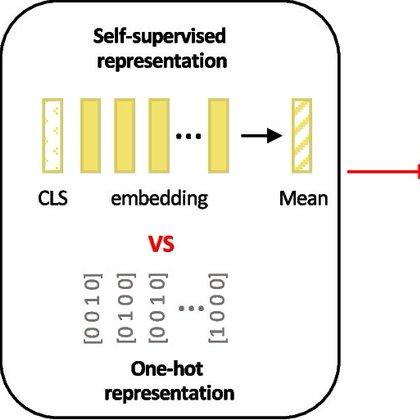
Do current genomic language models (pre-trained on whole genomes) learn a foundational understanding of biology in the non-coding region of human genomes? A new evaluation led by @AmberZqt suggests not yet! 1/N paper:
1
42
125
Thrilled to open-source the dataset behind our @NatMachIntell cover paper! 🧬 The ChatNT training data is now open-source on @huggingface. It's the first large-scale dataset for training conversational agents on biological sequences. A thread on what's inside 👇
1
8
16
🧬 Genomic DNNs can be trained to learn a lot of different aspects of gene regulation, but they're not perfect and we don't know which predictions are reliable and which ones aren't. We introduce DEGU: Uncertainty-aware Genomic Deep Learning with Knowledge Distillation. 1/n
1
20
84
Excited to share HIPPO (Histopathology Interventions of Patches for Predictive Outcomes)! HIPPO is a perturbation-based post hoc explanation tool interprets weakly supervised models for digital pathology. 1/N Work led by @JakubKaczmarzyk.
2
8
31
Excited to sponsor and participate in the Machine Learning for Computational Biology Conference #MLCB2024, kicking off today in Seattle! Watch @thomas_pierrot explain how InstaDeep is advancing Generative AI for Genomics! 🧬 📽️ Live here: https://t.co/irheoO0HuO
0
7
15
In genomic deep learning, the trends right now are to build bigger models that consider longer sequence contexts. While predictions are more powerful, their scale makes them difficult to interpret. To address this gap, we have developed CREME. Paper: https://t.co/ncQaFRjcC6 1/N
biorxiv.org
The rise of large-scale, sequence-based deep neural networks (DNNs) for predicting gene expression has introduced challenges in their evaluation and interpretation. Current evaluations align DNN...
1
40
125
Very excited to be at SysBio2024. Shoot me a message or just come chat with me about deep learning in regulatory genomics! #cshlsysbio
0
6
17
Do current genomic language models (pre-trained on whole genomes) learn a foundational understanding of biology in the non-coding region of human genomes? A new evaluation led by @AmberZqt suggests not yet! 1/N paper:
biorxiv.org
The emergence of genomic language models (gLMs) offers an unsupervised approach to learning a wide diversity of cis -regulatory patterns in the non-coding genome without requiring labels of functio...
10
58
238
Excited for #MLCB2023! Check out talk by @ToneyanSh on uncovering higher-order CRE interactions from large-scale DNNs and 2 posters: revisiting inits by @ckochath and @AmberZqt and new attribution method using domain-inspired surrogate models by @EESeitz! https://t.co/dC8slzIgkZ
2
8
33
Excited to share new work on "Interpreting cis-regulatory mechanisms from genomic deep neural networks using surrogate models” led by @EESeitz, jointly advised by me and @jbkinney and in collab with @TheDMMcC Paper: https://t.co/dC8slzHIvr Docs: https://t.co/R0AF6ZgdAr
5
40
110
Excited to share new work with @ToneyanSh! CREME (Cis-Regulatory Element Model Explanations), a suite of in silico perturbation experiments to uncover the rules of gene regulation learned by large-scale DNNs trained on functional genomics data. https://t.co/5glSrrtEew
1
26
92
Excited to share new work from my lab on "Evolution-inspired augmentations improve deep learning for regulatory genomics" Paper: https://t.co/uB1CwC8YjK Code: https://t.co/7OIkZgbE7e Analysis: https://t.co/UrRvBP8oWy 1/N
github.com
Contribute to p-koo/evoaug_analysis development by creating an account on GitHub.
3
15
66
New work led by Shushan Toneyan and Ziqi (Amber) Tang (@AmberZqt) on evaluating deep learning models for predicting epigenomic profiles. #RegulatoryGenomics 1/5 https://t.co/qp62ZmxOYB
3
33
162