Almut Eisele
@AlmutEisele
Followers
151
Following
150
Media
6
Statuses
60
Postdoctoral researcher @EPFL -transcriptional and epigenetic memory- lineage tracing- stem cells- single cells- multiomics
Joined December 2021
This is an incredible result! Allows you to find memory genes from a static snapshot of single cell analysis. Wow!🤯
Inheritable 'memory' genes drive #cancer drug tolerance, but their discovery in clinical samples is a major challenge. Our new theory, moving away from Luria-Delbruck, detects them from a *single* scRNA-seq dataset! With #ArchishmanRaju led by @suvranil_1
3
4
37
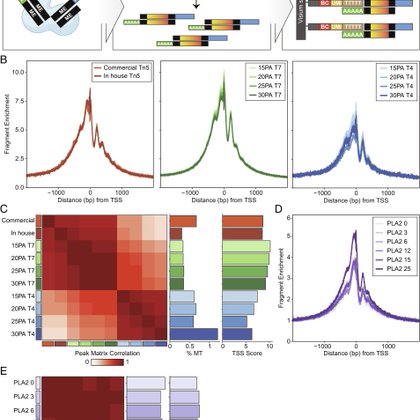
Happy to share our paper describing SPACE-seq - a new approach to spatial multiomics and lineage tracing - out today in @PNASNews! 🧵(1/5) https://t.co/bzTm0fqasH
pnas.org
Spatial epigenomics and multiomics can provide fine-grained insights into cellular states but their widespread adoption is limited by the requireme...
5
30
144
@EMBO workshop on Single cell Lineage Tracing! Sept 18-21 in Sant Feliu, Girona, Spain. Registration is OPEN!!!!! Tons of opportunities for talks are still available (including full length ones). https://t.co/5zyDYe4ZaU
#EMBOlineagetracing
meetings.embo.org
Recent advances in high-resolution lineage tracing technologies and single-cell genomics are allowing researchers to quantify cell state dynamics at unprecedented scales. Across cancer and immunity, …
0
6
19
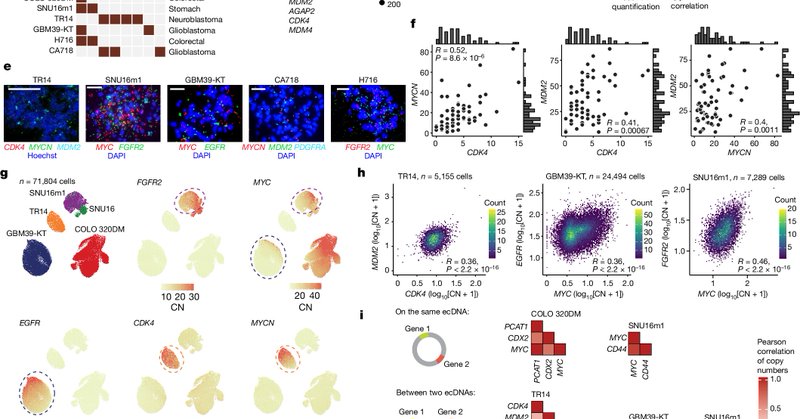
Cancer cells often harbor oncogenes outside chromosomes on extrachromosomal DNA (ecDNA), which is unevenly inherited by dividing cells. Today we report in @Nature that collectives of ecDNAs are inherited together by dividing cancer cells. 1/ https://t.co/scC2sc61Gh
nature.com
Nature - Cooperative species of extrachromosomal DNAs are coordinately inherited through mitotic co-segregation.
8
105
384
Same cell type, same trans environment, same sequence—different expression pattern. Locking in of stochastic differences into epigenetic memory. I just think this is so cool!
Through generation of resistant colonies from a clonal population of cells containing this dual reporter construct, we found that this memory encoding occurs in cis. (6/9)
7
17
94
Got out of the nucleus! Tested ~30,000 tiles from human RNA-binding proteins, found lots of domains that downregulate RNA&built an inducible synthetic RNA-binding protein for keeping target RNA at constant intermediate levels: https://t.co/g6d7XEZOlY So many follow-up projects!
biorxiv.org
RNA regulation plays an integral role in tuning gene expression and is controlled by thousands of RNA-binding proteins (RBPs). We develop and use a high-throughput recruitment assay (HT-RNA-Recruit)...
Do human RNA-binding proteins have modular regulatory domains that downregulate RNA lifetimes? Our preprint @BintuLab studying this question is now out on bioRxiv, where we indeed identify small domains in RBPs that potently induce RNA degradation! https://t.co/nOsgx1a3gA (1/9)
1
25
104
📢 To maintain proteome homeostasis, cells need to coordinate the production and elimination of proteins. How does it work? Check out our new preprint: https://t.co/PlaMkmUFgu A 🧵
biorxiv.org
Protein synthesis and decay rates must be tightly coordinated to ensure proteome homeostasis. How this coordination is established is unknown. Here we use quantitative live cell imaging combined with...
2
16
76
Hello Berlin students, we are looking for a motivated student assistant to support our research team at @berlinnovation and @BIMSB_MDC: https://t.co/4mnSJuUYKz. Please apply by July 20! What we do: https://t.co/6twsZ2hCms and https://t.co/uD1ZdHwF99 Retweets welcome 🙂
1
24
29
Second postdoc opening on our #ERC synergy project with @gagneurlab and @KevinVerstrepen . If you want to do RNA biology at scale combining genomics, synthetic biology and Machine learning. Join us at @karolinskainst @scilifelab Deadline 2024-06-15 https://t.co/cJL6dv0oDc
ki.varbi.com
Do you want to contribute to top quality medical research? The Department of Microbiology, Tumor and Cell Biology (MTC) at Karolinska Institutet conducts research and teaching within immunology, infec
2
12
20
@davidsuter_epfl @MTarbier @vpelech Many thanks also to the EPFL platforms involved! Gene Expression Core Facility, @BastienMangeat @Cytometry_EPFL, Biomolecular Screening Facility. And finally thanks to the feedback from numerous people at EPFL and beyond @GioeleLaManno @BartDeplancke
0
0
2
Massive thanks to everyone involved! In particular my supervisor @davidsuter_epfl and my collaborator @MTarbier and @vpelech, as well as the fantastic student Alexia Dormann.
1
0
0
7) GEMLI is available as R package on GitHub: https://t.co/RYx1mnPUVD Check it out! We are happy to help.
github.com
Contribute to UPSUTER/GEMLI development by creating an account on GitHub.
1
0
0
6) GEMLI fills a methodological gap left by other lineage tracing techniques as it can identify closely-related cells in human samples! This can also help to refine lineage trees generated by other techniques.
1
0
0
5) …further, GEMLI is great to study cell fate decisions by identifying closely-related cells undergoing various differentiation processes. In human breast cancer GEMLI mapped lineages making the transition to invasiveness- something no other technique can achieve!
2
0
1
4) GEMLI can reconstruct individual multicellular structures as individual organoids, crypts or small metastases ..
1
0
0
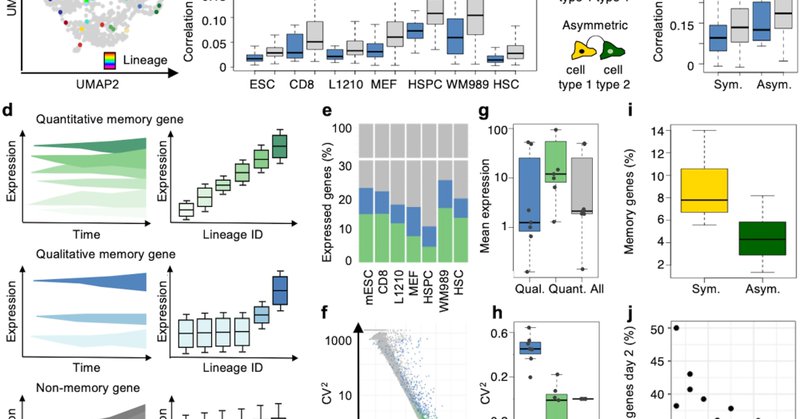
3) Leveraging these “memory genes” we developed GEMLI – Gene Expression Memory-based Lineage Inference, a computational tool to annotate lineages in scRNA-seq datasets solely based on gene-expression.
1
0
0
2) We could show that in any cell type we studied, from fibroblasts, to hematopoietic cells, immune cells, breast- prostate- or leukemic- cancer cells, and embryonic stem cells, thousands of “memory genes” maintain their gene expression levels over cell divisions.
1
0
0
1) The lineage tracing techniques with scRNA-seq readout today rely mainly on genetic marks. This restricts their use and time resolution. Could we use gene expression alone to identify lineages in scRNA-seq data? Yes we can! ..
1
0
0
Do you want to annotate lineages in your scRNA-seq datasets solely using gene expression? GEMLI is for you! Work from my postdoc with and thanks to @davidsuter_epfl @EPFL in collaboration with @MTarbier and @pelechanolab from @SciLifelab
nature.com
Nature Communications - Combining experimental lineage tracing with single cell transcriptomics is technically demanding. Here, authors present GEMLI, a computational tool to annotate cell lineages...
2
6
28