Kyle Tretina, Ph.D.
@AllThingsApx
Followers
4K
Following
16K
Media
632
Statuses
8K
Follow for AI in Digital Biology and Drug Discovery @NVIDIA, ex Insilico Medicine, ex Yale, PhD UMaryland, views are mine, DM for collabs
Boston, MA
Joined November 2013
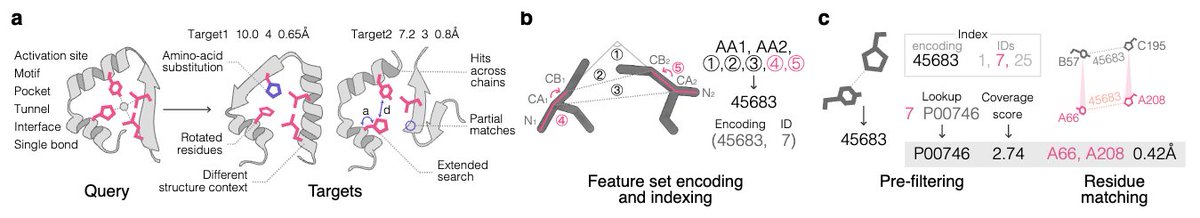
🧬Motifs are the new BLAST. Folddisco scans 53M AlphaFold structures for catalytic, binding or allosteric patterns in seconds🔎. 18× quicker, 3.5× smaller, hits in seconds🌌
5
50
348
RT @genophoria: We put a blog post together to tell you about all that went behind the scenes for @arcinstitute's Virtual Cell Challenge: (….
0
32
0
RT @gdb: Our custom LLM, gpt-4b micro, has helped achieve an advance in biology. It designed novel variants of the Nobel-winning Yamanaka….
0
173
0
RT @ikalvet: RFdiffusion2 is now live!. You can now design proteins, and in particular enzymes from just partially….
0
95
0
RT @EpochAIResearch: What's the best model you can run on a single consumer GPU?. We've updated last week's Data Insight with 3 additional….
0
26
0
RT @NikoMcCarty: I'm (slowly) writing the book I've been thinking about for the last 3+ years. Nothing official yet, but I'm hoping to wri….
0
148
0
A new self driving lab out of NC State University:.
sciencedaily.com
A new leap in lab automation is shaking up how scientists discover materials. By switching from slow, traditional methods to real-time, dynamic chemical experiments, researchers have created a...
0
0
1
Read the preprint:. Quantum-Boosted High-Fidelity Deep Learning.
arxiv.org
A fundamental limitation of probabilistic deep learning is its predominant reliance on Gaussian priors. This simplistic assumption prevents models from accurately capturing the complex,...
1
1
1
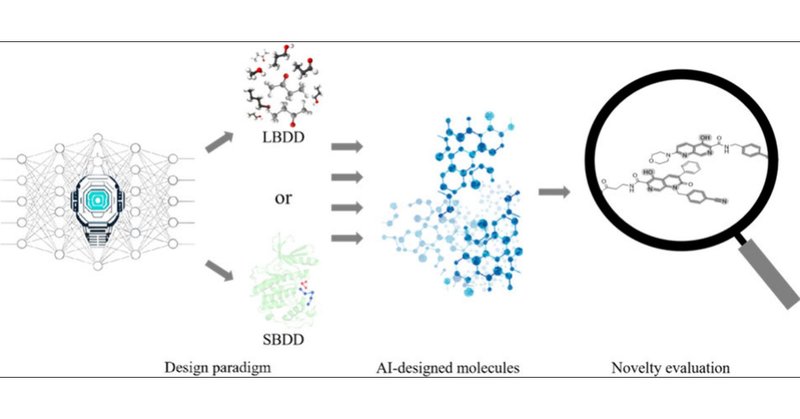
The paper:. AI-Designed Molecules in Drug Discovery, Structural Novelty Evaluation, and Implications.
pubs.acs.org
Achieving structural novelty in drug discovery remains a critical challenge. Artificial intelligence (AI) has demonstrated remarkable potential in deciphering the complex relationships between...
1
0
0
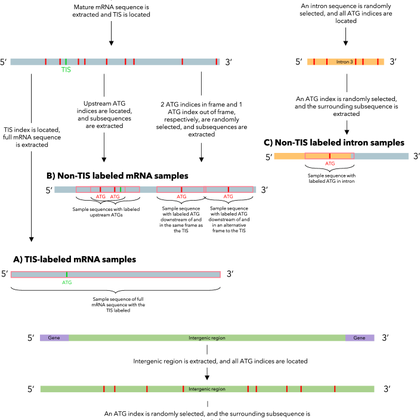
Paper:. NetStart 2.0: prediction of eukaryotic translation initiation sites using a protein language model.
bmcbioinformatics.biomedcentral.com
Background Accurate identification of translation initiation sites is essential for the proper translation of mRNA into functional proteins. In eukaryotes, the choice of the translation initiation...
0
0
3
RT @PossuHuangLab: We have a new collection of protein structure generative models which we call Protpardelle-1c. It builds on the original….
0
28
0