Wenliang Wang
@wenliang272
Followers
152
Following
633
Media
14
Statuses
74
Postdoc in Ecker lab at Salk. Broadly interested in (epi)genomics, immunology, neuroscience, and everything in between. Marathon and triathlon enthusiast.
San Diego, CA
Joined August 2012
Paper: https://t.co/HExCfg6TAf DNA methylation genome browser: https://t.co/Rc95suzxmj
#Alzheimers #Epigenomics #SingleCell #multiomic
biorxiv.org
Alzheimer’s disease (AD) is the most common neurodegenerative disorder, yet the molecular mechanisms underlying its region– and cell-type-specific pathogenesis remain poorly defined. Here, we...
0
2
6
9/ It’s the Year of Alzheimer’s at @salkinstitute 🧠 Proud to share our new single-cell multi-omic study of AD, alongside many exciting studies from colleagues — all driving fresh insight into mechanisms & therapies.
1
1
4
8/ Big thanks to our amazing collaborators across multiple institutions — and especially to my mentors @JoeEcker, Bradley Hyman and Bing Ren for their guidance and support throughout this project. 🙏
1
1
2
7/ Global epigenomic change and transcriptional repression in stressed cell state Stressed cell state has higher boundary density, associated with more short-range contact, and transcription repression.
1
1
1
6/ Key finding 2 - Genomic deletions near telomeres in AD-associated cell states 🧬 These states span neurons & glia, marked recurrent telomere-proximal deletions.
1
1
1
5/ Key finding 1 - opposing association between VC and TC/PFC 🧬 Consistent with the vulnerability of AD, almost every feature is opposingly associated with AD in VC and TC, with PFC showing intermediate profile.
1
1
1
4/ 3D genome in AD 🧩 We see widespread reorganization: disrupted contact–distance patterns, shifted compartments, more TAD boundaries, and increased chromatin loops — highlighting large-scale 3D genome remodeling in AD.
1
1
1
3/ DNA methylation in AD 🧬 Temporal cortex → genome-wide hypermethylation Visual cortex → genome-wide hypomethylation in many cell types Prefrontal cortex → intermediate We defined two recurrent cell states: 🔹 Homeostatic (normal-like) 🔹 Stressed (epigenomically disrupted)
1
1
1
2/ AD is the most common neurodegenerative disorder, but why some brain regions are more vulnerable than others has remained unclear. We profiled temporal cortex (TC), prefrontal cortex (PFC), and visual cortex (VC) — spanning severe to relatively spared regions.
1
1
2
🧵1/ Excited to share our new paper: "Brain Region-Specific Epigenomic Reorganization and Altered Cell States in Alzheimer’s Disease" We built a single-cell multi-omic atlas (DNA methylation + 3D genome) from AD and control brains. Paper:
biorxiv.org
Alzheimer’s disease (AD) is the most common neurodegenerative disorder, yet the molecular mechanisms underlying its region– and cell-type-specific pathogenesis remain poorly defined. Here, we...
2
18
50
Genetics and Environment Distinctively Shape the Human Immune Cell Epigenome
biorxiv.org
The epigenomic landscape of human immune cells is dynamically shaped by both genetic factors and environmental exposures. However, the relative contributions of these elements are still not fully...
0
2
5
So disappointing! Can't believe this happened in 2024 at @MIT
@NeurIPSConf
As alumni I'm sad to see a mit professor publicly shaming chinese students in a top conference without citing credible sources. The more serious question is: did she make it up? I dont see any sensible person would make such a confession. @MIT @NeurIPSConf
0
0
3
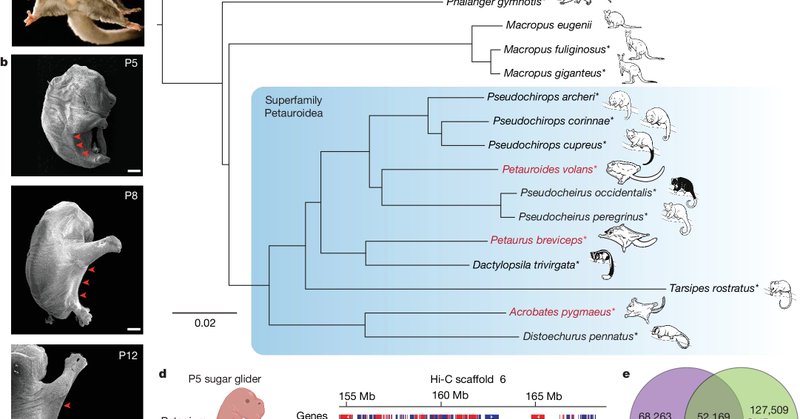
Mark. Another example combining evolutionary and functional genomics.
nature.com
Nature - Patagia—the mammalian gliding membrane—repeatedly originated through a process of convergent genomic evolution, whereby the regulation of Emx2 was altered by distinct...
0
0
4
Great resource for writing the application package from the Dr. Ross-Ibarra's lab
Your regular reminder, as job season is upon us, that there are a ton of successful job applications (industry, PUI, and lots of research university) available here: https://t.co/v0NChnm9Mr. Please consider contributing your own if you've recently landed a gig!
0
0
0
Thrilled to share that I will start my lab at @arcinstitute as a Science Fellow, joining this amazing community of diverse and creative scientists. Looking forward to the synergy between us to quantitatively model biological systems and link the knowledge to human diseases.
@zhou_jingtian joins as our first Science Fellow. Jingtian is a recent PhD graduate in Bioinformatics & Systems Biology from @UCSD and a former Salk Institute scientist. His pioneering work has developed new algorithms and insights into the epigenetic basis of brain development
10
6
78
Have been using this tool for a while, happy to see it out in @NatureBiotech. Congratulations! By the way, will you guys consider single-cell methylation data for somatic mutation?
Very excited to see our study presenting SComatic out in @NatureBiotech! Huge congrats to @fran_muyas and all co-authors! paper: https://t.co/IpgP7VusVD briefing: https://t.co/VUnLDg2sej news: https://t.co/o7I5tB6MjO thread👇
1
0
2
Finally, we would like to acknowledge the immense contributions of our donors, sample collection team, and data generation team, including Anna, Rosa, Cesar, @SongHaili and many others. This study would not have been possible without their tremendous efforts and dedication.
0
1
2
We express our gratitude to @DARPA for their generous support, as well as our collaborators @WJGreenleaf @EpigenomeI Bei Wei, Irem for the discussions and constructive suggestions. Lastly, we extend a heartfelt appreciation to our mentor, @JoeEcker, for his exceptional guidance.
1
0
2