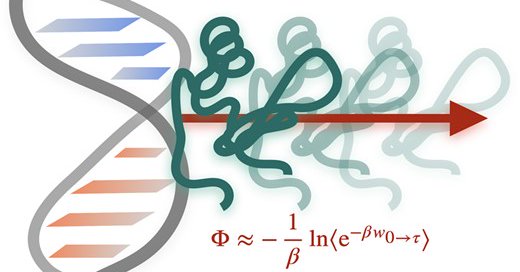Thor van Heesch
@thorvanheesch
Followers
75
Following
1K
Media
0
Statuses
50
PhD Candidate @CompChemUvA Interested in enhanced sampling methods for biomolecular systems...
Joined February 2020
Super excited to share together with @jocelyne_vreede and Peter Bolhuis my first research paper published in Nucleic Acids Research! We have developed a molecular simulation approach for predicting dissociation free energies of protein-DNA complexes:
1
2
21
Monte Carlo with expanded ensembles are powerful methods to compute adsorption and diffusion processes in nanoporous materials. In @JChemPhys , we present RASPA3, the new version of the RASPA2 code. (1/8) https://t.co/oiz69FN3D9
1
24
59
~2 weeks left to apply! Thanks for your interest & RTs.
🚨 Hiring! Looking for a motivated #PhD student to work with me on molecular #simulations and #machinelearning to predict #nanoplastic risks to #protein function. Further details: https://t.co/CAUBAWyHyR RT appreciated #compchem #softmatter @jobRxiv @HimsUva @UvA_Science
0
6
7
🚨 Hiring! Looking for a motivated #PhD student to work with me on molecular #simulations and #machinelearning to predict #nanoplastic risks to #protein function. Further details: https://t.co/CAUBAWyHyR RT appreciated #compchem #softmatter @jobRxiv @HimsUva @UvA_Science
0
18
36
🔬 Happy to announce our latest publication "Unlocking Novel Therapies: Cyclic Peptide Design for Amyloidogenic Targets through Synergies of Experiments, Simulations, and Machine Learning." 📚 #AmyloidogenicTargets #ML #CyclicPeptideDesign
https://t.co/DEe6xUyIIY
0
3
18
For more in-depth insights and detailed methodology, check out our full paper in @NAR_Open
academic.oup.com
Abstract. Sequence-specific protein–DNA interactions are crucial in processes such as DNA organization, gene regulation and DNA replication. Obtaining deta
0
0
0
Our method's versatility extends beyond minor groove binders like H-NS. It's equally effective for major groove binding proteins. We applied our technique to a major groove binder, the ETS domain of the PU.1 TF. Again, we were able to identify the consensus sequence correctly!
1
0
1
We successfully applied this technique to analyze the Histone-like Nucleoid Structuring (H-NS) protein, a notable minor groove binder. Our predictions align well with experimental data, revealing H-NS’s sequence-specific binding to AT-rich DNA
1
0
0
We employed molecular dynamics simulations, Jarzynski's equality, and a unique steering potential based on a contact map. This combination allows us to effectively calculate free energy differences in protein-DNA dissociation, distinguishing our approach from traditional methods
1
0
0
Our paper introduces a new method to quantify the sequence specificity of protein-DNA complexes, offering high-resolution insights into recognition mechanisms. This is crucial for understanding DNA organization, gene regulation, and DNA replication
1
0
0
Hello @IUPAC2023! Today at 10:20, during the #Chemistry & #AI session, I will present our work with @thorvanheesch & @BerndEnsing from @CompChemUvA @HimsUva @UvA_Science on finding molecular descriptors from (meta)stable-state data :)
0
1
37
I made a free addon for pixel art rendering with blender! full tutorial and download link: https://t.co/13zTlNVnH2
#blender #b3d #pixelart
32
611
3K
🚨Looking for 2 postdocs to join @UvA_Science @ai4science_lab 4 sustainable molecules & materials! Salt hydrates 4 thermal storage (with Shahidzadeh, Woutersen) Plant proteins 4 sustainable food (Bolhuis, van Hoof, Jabbari-Farouji, Quattrocchio, Schall) https://t.co/4yJ9Lbys2D
0
10
17
🚨 Hiring! Looking for a motivated PhD student to work with me on automated computational design of molecules & soft materials for sustainability & health. Further details: https://t.co/KpDneyBKYQ Retweets appreciated #compchem #softmatter @Chemjobber @jobRxiv
1
19
37
I'm after any molecular dynamics simulation data for some upcoming talks / videos. Can anyone help me out? I'm currently after ideally a simulation showing ions passing through an ion channel. Ideally lots of time steps, lots of atoms (waters, ions, EVERYTHING)
21
25
194
@lal_ashique sharing the details of his PhD project under the supervision of Peter Bolhuis and Evert Jan Meijer #compchem @ARCCBBC
Introducing our new PhD candidates: Nicolette Maaskant (@UniUtrecht), @lal_ashique (@UvA_Amsterdam) & Sven Weerdenburg (@tudelft). They introduce themselves and their research 👉 https://t.co/EREq0CraD9
0
1
4
0
4
17
A single neural network that predicts for protein-DNA complex structures, protein-RNA complex structures, and RNA tertiary structures. @minkbaek Ryan McHugh, Ivan Anishchenko, David Baker, Frank DiMaio @UWproteindesign
https://t.co/yzsATo48n4
2
95
383
"Language Models Can Teach Themselves to Program Better" This paper changed my thinking about what future langauge models will be good at, mostly in a really concerning way. Let's start with some context: [1/11]
41
615
3K
For my latest attempt at introducing proteins to students, I made a Google Colab Notebook that predicts proteins from a single sequence. I asked the students to tweak the sequence to get a helix or two helices or... (1/5) https://t.co/eRufxDAkWm
19
284
2K