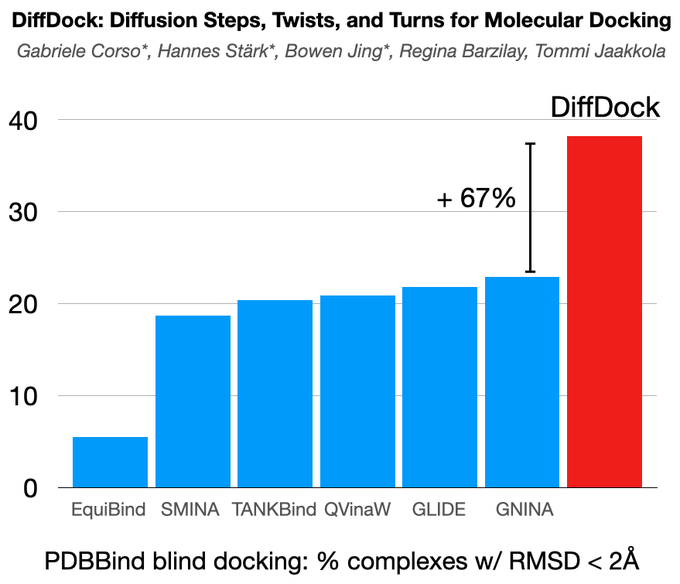
DiffDock is now on
@huggingface
Spaces with interactive visualization of the diffusion steps. Built using
@Gradio
and 3dmol.js and you can upload your own ligands as SMILES or sdf/mol2. Happy docking!
1/2
6
76
314
Replies
Props to the authors (
@GabriCorso
@HannesStaerk
et al.) that made the code available in contrast to most other papers for diffusion models in the bio space . Would love to add other models in the space as comparison. 2/2
0
1
17