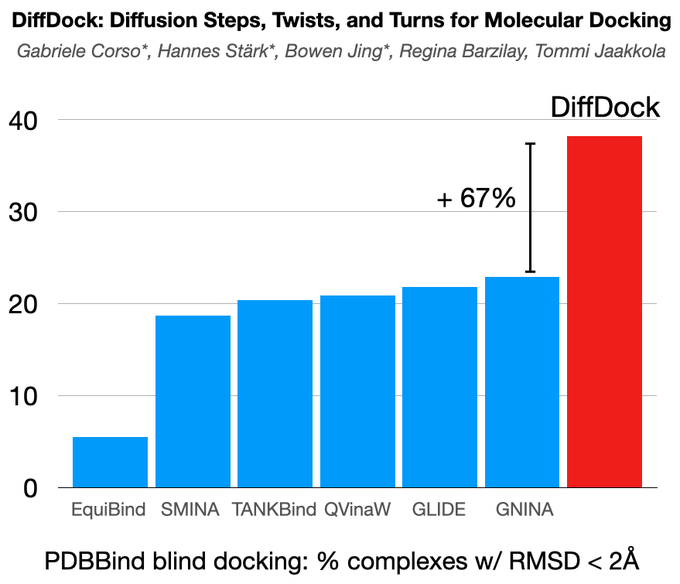
Excited to share DiffDock, new non-Euclidean diffusion for molecular docking! In PDBBind, standard benchmark, DiffDock outperforms by a huge margin (38% vs 23%) the previous state-of-the-art methods that were based on expensive search!
A thread! 👇
16
96
450
Replies
@HannesStaerk
@BarzilayRegina
Recent regression-based ML methods for docking showed strong speed-up but no significant accuracy improvements over traditional search-based approaches. We identify the problem in their objective functions and show how generative modeling aligns well with the docking task.
2
1
14
@HannesStaerk
@BarzilayRegina
We, therefore, develop DiffDock, a non-Euclidean diffusion model over the space of ligand poses for molecular docking. DiffDock defines a diffusion process over the position of the ligand relative to the protein, its orientation, and the torsion angles describing its conformation
1
1
11
@HannesStaerk
@BarzilayRegina
To efficiently train and run the diffusion model over this highly non-linear manifold, we map the elements of the manifold to a product space of T(3) x SO(3) x SO(2)^m groups corresponding to the translation, rotation, and torsion transformations.
1
0
6
@HannesStaerk
@BarzilayRegina
We achieve a new state-of-the-art 38% top-1 prediction with RMSD<2A on the PDBBind blind docking benchmark, considerably surpassing the previous best search-based (23%) and deep learning methods (20%). DiffDock also has faster runtimes than the previous state-of-the-art methods.
1
0
10
@HannesStaerk
@BarzilayRegina
We also train a confidence model that can rank poses by likelihood and predict the model's confidence in the generated poses. Experimentally, this confidence score provides high selective accuracy, reaching 83% on its most confident third of the previously unseen complexes.
1
0
7
@HannesStaerk
@BarzilayRegina
The project resulted from a great collaboration with
@HannesStaerk
(joint first), Bowen Jing (joint first),
@BarzilayRegina
and Tommi Jaakkola. Code and trained models are available at:
2
0
22
1
1
13
@GabriCorso
@HannesStaerk
@BarzilayRegina
Hi Gabriele. Great work ! Do you have a link or two to the performance of the “other” methods in pdbbind? Or at least the exact details of the test you did ? Thanks !
1
0
4
@adrian_roitberg
@HannesStaerk
@BarzilayRegina
We mainly follow the setup previously used in EquiBind and TANKBind, but we tried to put all the details on how the baselines are run in Appendix D.3 (see image below). Moreover, in the GitHub repository, we also provide many of the scripts to run and evaluate the baselines
1
0
1
@GabriCorso
@HannesStaerk
@BarzilayRegina
Thanks for the great work! I have a question about confidence score. What is the scale of the score? How can we interpret the score 1.56 vs 5.3? What do they tell and how can we use them?
1
0
0
@liwen_you
@HannesStaerk
@BarzilayRegina
It is the logit output (before soft max) of a binary classifier. Therefore, anything positive means the model thinks that there is more than 50% that the predicted pose is within 2A of the true one. However, note that the confidence model has not been calibrated on held out data.
1
1
0
@GabriCorso
@HannesStaerk
@BarzilayRegina
Hi, I'm trying to apply your model. What is the TOP 1 pose, the one with the highest or lowest confidence value? I tried to follow the videos, but in the end I didn't understand.
1
0
0
@GabriCorso
@HannesStaerk
@BarzilayRegina
Great work, I'm actually searching for gnina alternative to dock porphyrin conjugates for my master's thesis and will definitely check it out
0
0
2
@GabriCorso
@HannesStaerk
@BarzilayRegina
Catching up on this, the data and generative approach look exciting, have you tried a protein/protein docking set?
0
0
0